# PyPlaque
Python package for virus plaque analysis based on Plaque2.0
# Installation
See project's PyPi page [https://pypi.org/project/PyPlaque/](https://pypi.org/project/PyPlaque/)
```
pip install PyPlaque
```
# Local devloper installation
- Clone repo
- run `pip install -e .`
# Usage
```
from PyPlaque.specimen import PlaquesImageGray
from PyPlaque.phenotypes import Plaque
```
Here's a test example of how to analyze plaques in an image using PyPlaque. First let's load an example image:
```
from skimage.io import imsave, imread, imshow
img_url = 'https://raw.github.com/plaque2/matlab/master/Sample_B01_s1_w2.tif'
img = imread(img_url)
imshow(img)
```

Now let's detect plaques in this flourescence microscopy image and print out all their coordinmates:
```
plq_img = PlaquesImageGray('Sample_B01_s1_w2',img, threshold=0.25)
for plq in plq_img.get_plaques():
print(plq.centroid)
```
This will output following measurements:
```
(435.85833333333335, 1785.8416666666667)
(653.1464788732394, 1209.9647887323943)
(708.058912386707, 1251.6835347432025)
(728.976492712741, 1599.9073812881993)
(712.3316195372751, 1312.491002570694)
(747.0576576576576, 1174.0234234234233)
(752.5772277227722, 1119.0891089108911)
(782.8965517241379, 1151.510344827586)
...
```
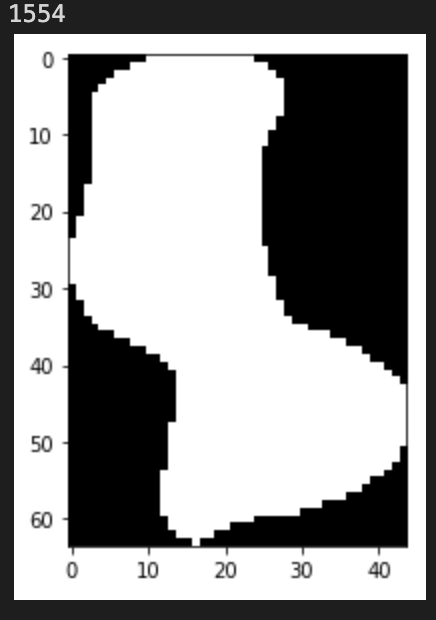
Each Plaque object containes a crop of individual virological plaque, as well as, measurements:
```
imshow(plq.mask)
print(plq.area)
```
___________
## Classes structure concept

## Specific classes
### Sepcimen
___________
**PlaquesMask class** designed to hold binary mask of multiple
plaque phenotypes.
_Arguments_:
name - (str, required) string, image sample name for identification
plaques_mask - (np.array, required) numpy array containing
binary mask of all virological plaque objects.
**get_palques method** returns a list of individual plaques
stored as binary numpy arrays.
_Arguments_:
min_are - (int, optional, default = 100) a cut-off value for plaque area
in px.
___________
**PlaqueImageGray class** designed to hold grayscale image data containing
multiple plaque phenotypes with a respective binary mask. The class inherits
from PlaquesMask.
_Additonal arguments_:
name - (str, required) string, image sample name for identification
image - (np.array, required) numpy array containing 2D grayscale image of
a virological plaque object, respective to the mask. Used, in case of
measuring properties of fluorescent plaque image.
plaques_mask - (np.array, optional, default None) numpy array containing
binary mask of all virological plaque objects.
threshold - (float between 0 and 1, optional, default None) fixed threshold
value for creating the binary mask.
sigma - (int, optional, default = 5) guassian blur sigma in pixels used by
the fixed thresholding approach.
Either mask or fixed threshold must be provided
___________
**PlaquesImageRGB class** designed to hold RGB image data containing
multiple plaque phenotypes with a respective binary mask.The class inherits
from PlaquesMask.
_Additonal arguments_:
name - (str, required) image sample name for identification
image - (np.array, required) 3D (red, green, blue) numpy array
containing image of a virological plaque object, respective to the mask.
Used, e.g. in case of measuring properties of crystal violet plaque image.
plaques_mask - (np.array, required) numpy array containing binary mask of all
virological plaque objects.
___________
**Class PlaquesWell** is aimed to contain a full well of a multititre plate.
_Arguments_:
row - (int or str, required) row id of the well.
column - (int or str, required) column id of the well.
well_image - (np.array, required) numpy array containing image of
the well.
well_mask - (np.array, required) numpy array containing binary mask of
the well.
___________
**PlateImage Class** is aimed to contain a full multititre plate image and
it's respective binary mask.
_Arguments_:
n_rows - (int, required) number of rows in the plate (usually lower than
the number of rows).
n_columns - (int, required) number of columns in the plate (usually higher than
the number of rows).
plate_image - (np.array, required) an image of individual wells of the
plate.
plate_mask - (np.array, required) a binary mask outlining individual wells of the
plate.
**get_wells method** returns a list of individual wells of the plate
stored as binary numpy arrays.
___________
### Phenotypes
___________
**Plaque** class is designed to hold a single virological plaque
phenotype as an object.
_Arguments_:
mask - (required, 2D numpy array) containing binary mask of a single
virological plaque object.
centroid - (float tuple, optional) contains x and y of the centroid of the
plaque object
bbox - (float tuple, optional) contains minr, minc, maxr, maxc of the
plaque object
___________
**FluorescencePlaque** conains plaque obtained from fluorescence image.
Class inherits from Plaque class and is also designed to hold a single
virological plaque phenotype.
_Additonal arguments_:
image - (required) numpy array containing grayscale image of a single
virological plaque object.
___________
**CrystalVioletPlaque** plaque obtained from crystal violet image. Class
inherits from Plaque class and is also designed to hold a single virological
plaque phenotype.
_Additonal arguments_:
image - (required) numpy array containing RGB or a graysacle image of a
single virological plaque object.
Raw data
{
"_id": null,
"home_page": "https://github.com/plaque2/PyPlaque",
"name": "PyPlaque",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "Virus,Plaque,Virology,Microscopy,Computer vision",
"author": "Plaque2.0 Team, AILS Institute, CASUS HZDR",
"author_email": "ayakimovich@ails.institute",
"download_url": "https://files.pythonhosted.org/packages/cf/18/3e7c873f16bef9762829892f71526aa5584fe7966f755ae9f56ce099f3ad/PyPlaque-0.1.0.tar.gz",
"platform": null,
"description": "# PyPlaque\nPython package for virus plaque analysis based on Plaque2.0\n\n# Installation\n\nSee project's PyPi page [https://pypi.org/project/PyPlaque/](https://pypi.org/project/PyPlaque/)\n\n```\npip install PyPlaque\n```\n\n# Local devloper installation\n\n- Clone repo\n- run `pip install -e .`\n\n# Usage\n\n```\nfrom PyPlaque.specimen import PlaquesImageGray\nfrom PyPlaque.phenotypes import Plaque\n```\n\nHere's a test example of how to analyze plaques in an image using PyPlaque. First let's load an example image:\n\n```\nfrom skimage.io import imsave, imread, imshow\n\nimg_url = 'https://raw.github.com/plaque2/matlab/master/Sample_B01_s1_w2.tif'\nimg = imread(img_url)\nimshow(img)\n```\n\n\n\nNow let's detect plaques in this flourescence microscopy image and print out all their coordinmates:\n\n```\nplq_img = PlaquesImageGray('Sample_B01_s1_w2',img, threshold=0.25)\nfor plq in plq_img.get_plaques():\n print(plq.centroid)\n```\n\nThis will output following measurements:\n\n```\n(435.85833333333335, 1785.8416666666667)\n(653.1464788732394, 1209.9647887323943)\n(708.058912386707, 1251.6835347432025)\n(728.976492712741, 1599.9073812881993)\n(712.3316195372751, 1312.491002570694)\n(747.0576576576576, 1174.0234234234233)\n(752.5772277227722, 1119.0891089108911)\n(782.8965517241379, 1151.510344827586)\n\n...\n```\n\nEach Plaque object containes a crop of individual virological plaque, as well as, measurements:\n\n```\nimshow(plq.mask)\nprint(plq.area)\n```\n\n\n\n___________\n\n## Classes structure concept\n\n\n\n\n## Specific classes\n\n### Sepcimen\n\n___________\n\n**PlaquesMask class** designed to hold binary mask of multiple\nplaque phenotypes.\n\n_Arguments_:\n\nname - (str, required) string, image sample name for identification\n\nplaques_mask - (np.array, required) numpy array containing\nbinary mask of all virological plaque objects.\n\n**get_palques method** returns a list of individual plaques\nstored as binary numpy arrays.\n\n_Arguments_:\n\nmin_are - (int, optional, default = 100) a cut-off value for plaque area\nin px.\n\n___________\n\n**PlaqueImageGray class** designed to hold grayscale image data containing\nmultiple plaque phenotypes with a respective binary mask. The class inherits\nfrom PlaquesMask.\n\n_Additonal arguments_:\n\nname - (str, required) string, image sample name for identification\n\nimage - (np.array, required) numpy array containing 2D grayscale image of\na virological plaque object, respective to the mask. Used, in case of\nmeasuring properties of fluorescent plaque image.\n\nplaques_mask - (np.array, optional, default None) numpy array containing\nbinary mask of all virological plaque objects.\n\nthreshold - (float between 0 and 1, optional, default None) fixed threshold\nvalue for creating the binary mask.\n\nsigma - (int, optional, default = 5) guassian blur sigma in pixels used by\nthe fixed thresholding approach.\n\nEither mask or fixed threshold must be provided\n___________\n\n**PlaquesImageRGB class** designed to hold RGB image data containing\nmultiple plaque phenotypes with a respective binary mask.The class inherits\nfrom PlaquesMask.\n\n_Additonal arguments_:\n\nname - (str, required) image sample name for identification\n\nimage - (np.array, required) 3D (red, green, blue) numpy array\ncontaining image of a virological plaque object, respective to the mask.\nUsed, e.g. in case of measuring properties of crystal violet plaque image.\n\nplaques_mask - (np.array, required) numpy array containing binary mask of all\nvirological plaque objects.\n\n___________\n\n**Class PlaquesWell** is aimed to contain a full well of a multititre plate.\n\n_Arguments_:\n\nrow - (int or str, required) row id of the well.\n\ncolumn - (int or str, required) column id of the well.\n\nwell_image - (np.array, required) numpy array containing image of\nthe well.\n\nwell_mask - (np.array, required) numpy array containing binary mask of\nthe well.\n\n\n___________\n\n**PlateImage Class** is aimed to contain a full multititre plate image and\nit's respective binary mask.\n\n_Arguments_:\n\nn_rows - (int, required) number of rows in the plate (usually lower than\nthe number of rows).\n\nn_columns - (int, required) number of columns in the plate (usually higher than\nthe number of rows).\n\nplate_image - (np.array, required) an image of individual wells of the\nplate.\n\nplate_mask - (np.array, required) a binary mask outlining individual wells of the\nplate.\n\n**get_wells method** returns a list of individual wells of the plate\nstored as binary numpy arrays.\n___________\n\n### Phenotypes\n\n___________\n\n**Plaque** class is designed to hold a single virological plaque\nphenotype as an object.\n\n_Arguments_:\n\nmask - (required, 2D numpy array) containing binary mask of a single\nvirological plaque object.\n\ncentroid - (float tuple, optional) contains x and y of the centroid of the\nplaque object\n\nbbox - (float tuple, optional) contains minr, minc, maxr, maxc of the\nplaque object\n\n___________\n\n**FluorescencePlaque** conains plaque obtained from fluorescence image.\nClass inherits from Plaque class and is also designed to hold a single\nvirological plaque phenotype.\n\n_Additonal arguments_:\n\nimage - (required) numpy array containing grayscale image of a single\nvirological plaque object.\n\n___________\n\n**CrystalVioletPlaque** plaque obtained from crystal violet image. Class\ninherits from Plaque class and is also designed to hold a single virological\nplaque phenotype.\n\n_Additonal arguments_:\n\nimage - (required) numpy array containing RGB or a graysacle image of a\nsingle virological plaque object.\n",
"bugtrack_url": null,
"license": "GPLv3",
"summary": "Python package for virus plaque analysis based on Plaque2.0",
"version": "0.1.0",
"project_urls": {
"Download": "https://github.com/plaque2/PyPlaque/archive/v0.1.0.tar.gz",
"Homepage": "https://github.com/plaque2/PyPlaque"
},
"split_keywords": [
"virus",
"plaque",
"virology",
"microscopy",
"computer vision"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "5a01ac948e8747525747833b627123ddf50df5af78ba944d74e42e3f7d2826b4",
"md5": "d2bcab7e36168fb2b041a8e4646637d1",
"sha256": "8da3684a510677c8d9ed9548a6e437061e0d94e257ab3d795ecd371cc47ed8ae"
},
"downloads": -1,
"filename": "PyPlaque-0.1.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "d2bcab7e36168fb2b041a8e4646637d1",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": null,
"size": 23585,
"upload_time": "2024-02-27T14:51:07",
"upload_time_iso_8601": "2024-02-27T14:51:07.578056Z",
"url": "https://files.pythonhosted.org/packages/5a/01/ac948e8747525747833b627123ddf50df5af78ba944d74e42e3f7d2826b4/PyPlaque-0.1.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "cf183e7c873f16bef9762829892f71526aa5584fe7966f755ae9f56ce099f3ad",
"md5": "8ac95d7973364a78223ff4b133a5da2e",
"sha256": "2080c2e6fdac2423b79a7eed7d54651f60817ae0185a28d6d7b3fe69d6331b4f"
},
"downloads": -1,
"filename": "PyPlaque-0.1.0.tar.gz",
"has_sig": false,
"md5_digest": "8ac95d7973364a78223ff4b133a5da2e",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 21975,
"upload_time": "2024-02-27T14:51:10",
"upload_time_iso_8601": "2024-02-27T14:51:10.919464Z",
"url": "https://files.pythonhosted.org/packages/cf/18/3e7c873f16bef9762829892f71526aa5584fe7966f755ae9f56ce099f3ad/PyPlaque-0.1.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-02-27 14:51:10",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "plaque2",
"github_project": "PyPlaque",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [
{
"name": "numpy",
"specs": []
},
{
"name": "scikit-image",
"specs": []
},
{
"name": "opencv-python",
"specs": []
},
{
"name": "matplotlib",
"specs": []
},
{
"name": "Pillow",
"specs": []
},
{
"name": "scipy",
"specs": []
},
{
"name": "pytest",
"specs": []
}
],
"lcname": "pyplaque"
}
