<img src="docs/source/images/divbrowse_logo.png" width="600">
<br />
[](https://pypi.org/project/divbrowse/)
[](https://hub.docker.com/r/ipkbit/divbrowse)

[](https://doi.org/10.1093/gigascience/giad025)
[](https://divbrowse.readthedocs.io/?badge=latest)
[](https://badge.fury.io/py/divbrowse)
[](https://pypi.org/project/divbrowse/)
[](https://libraries.io/pypi/divbrowse)

<br />
**Website:** https://divbrowse.ipk-gatersleben.de
**Documentation:** https://divbrowse.readthedocs.io
**Paper:** https://doi.org/10.1093/gigascience/giad025
<hr />
**Table of contents:**
- [About DivBrowse](#about-divbrowse)
- [Try out DivBrowse](#try-out-divbrowse)
- [Screenshots](#screenshots)
- [Usage workflow concept](#usage-workflow-concept)
- [Architecture](#architecture)
<br />
## About DivBrowse
DivBrowse is a web application for interactive exploration and analysis of very large SNP matrices.
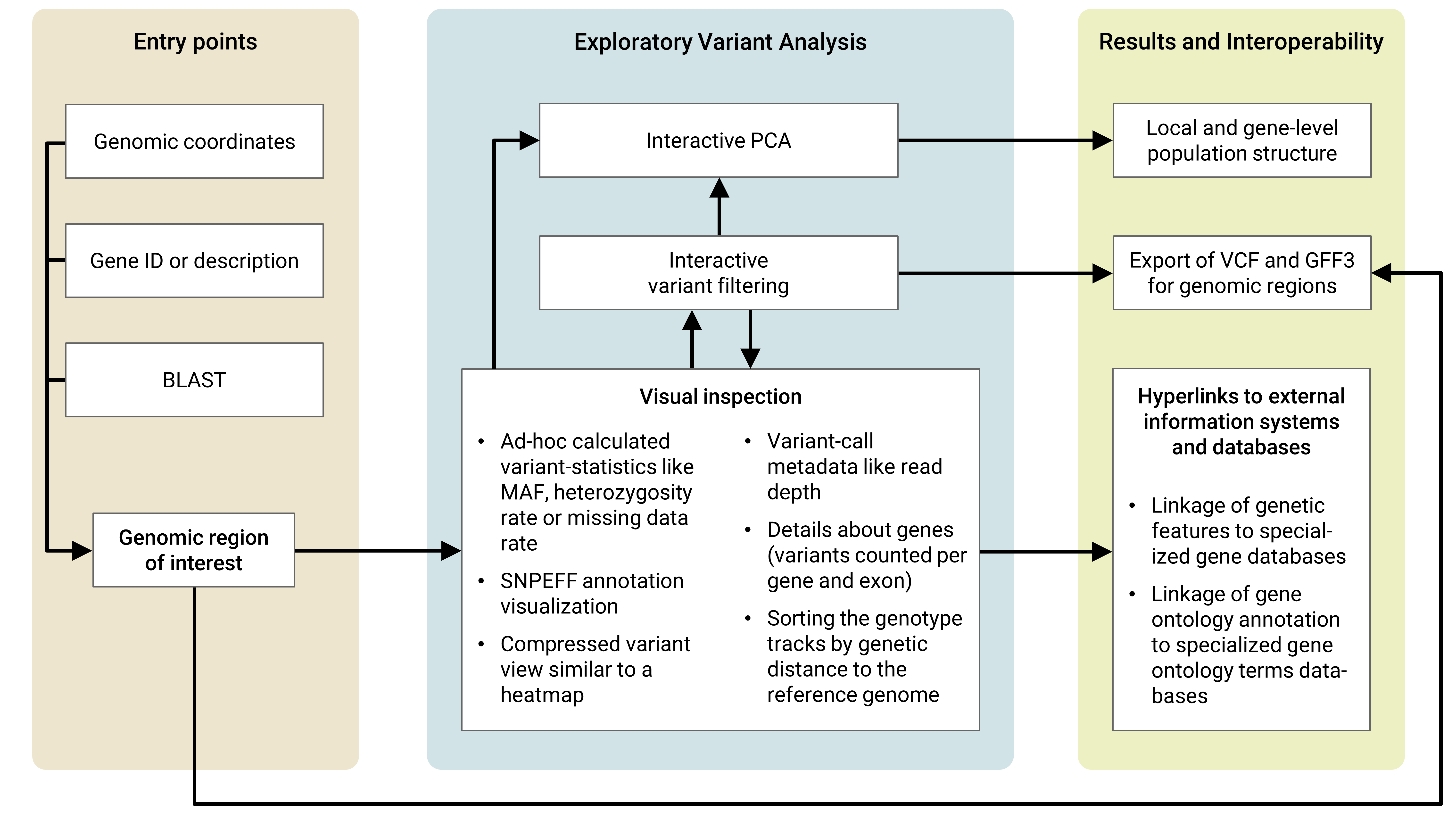
It offers a novel approach for interactive visualization and analysis of genomic diversity data and optionally also gene annotation data. The use of standard file formats for data input supports interoperability and seamless deployment of application instances based on established bioinformatics pipelines. The possible integration into 3rd-party web applications supports interoperability and reusability.
The integrated ad-hoc calculation of variant summary statistics and principal component analysis enables the user to perform interactive analysis of population structure for single genetic features like genes, exons and promoter regions. Data interoperability is achieved by the possibility to export genomic diversity data for genomic regions of interest in standardized VCF files.
## Try out DivBrowse
If you want to test DivBrowse please visit the demo instances listed here:
https://divbrowse.ipk-gatersleben.de/#demo-instances
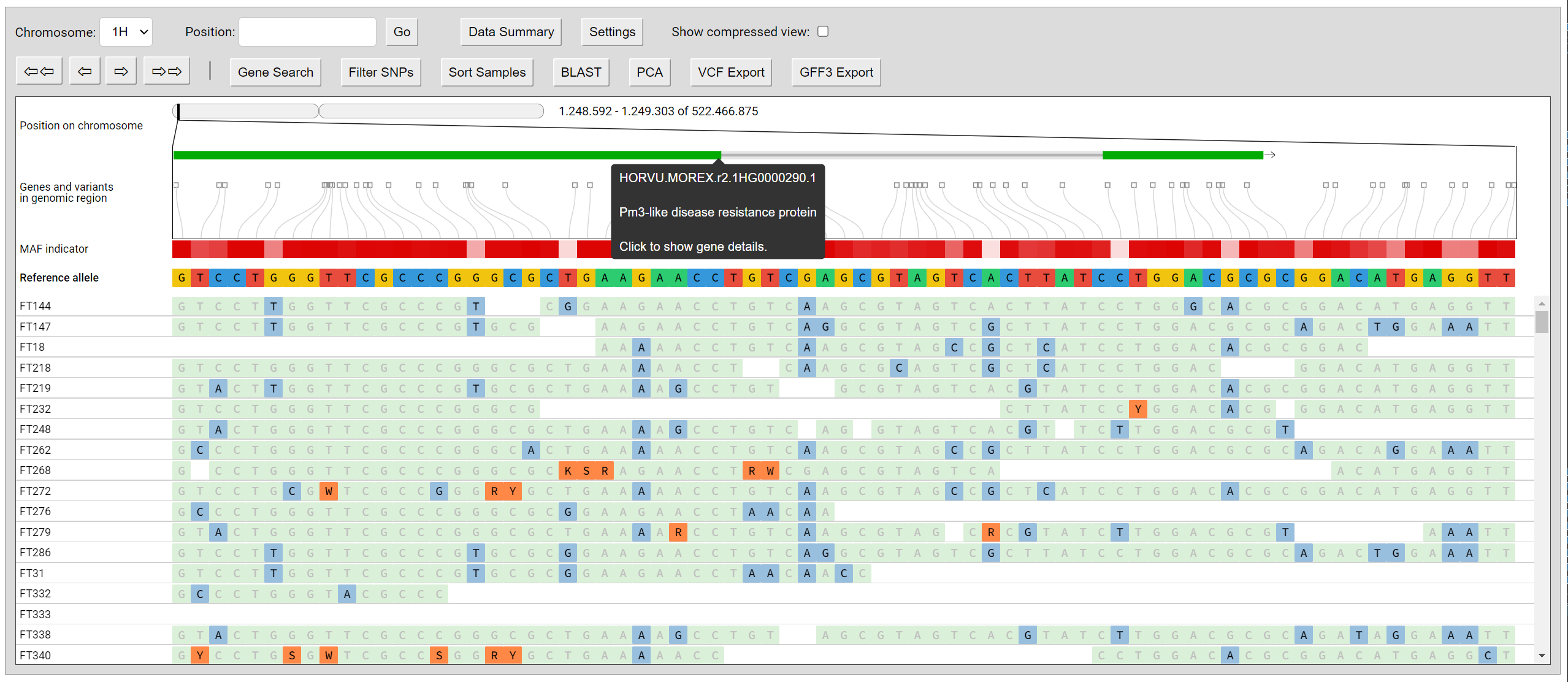
## Screenshots
## Usage workflow concept
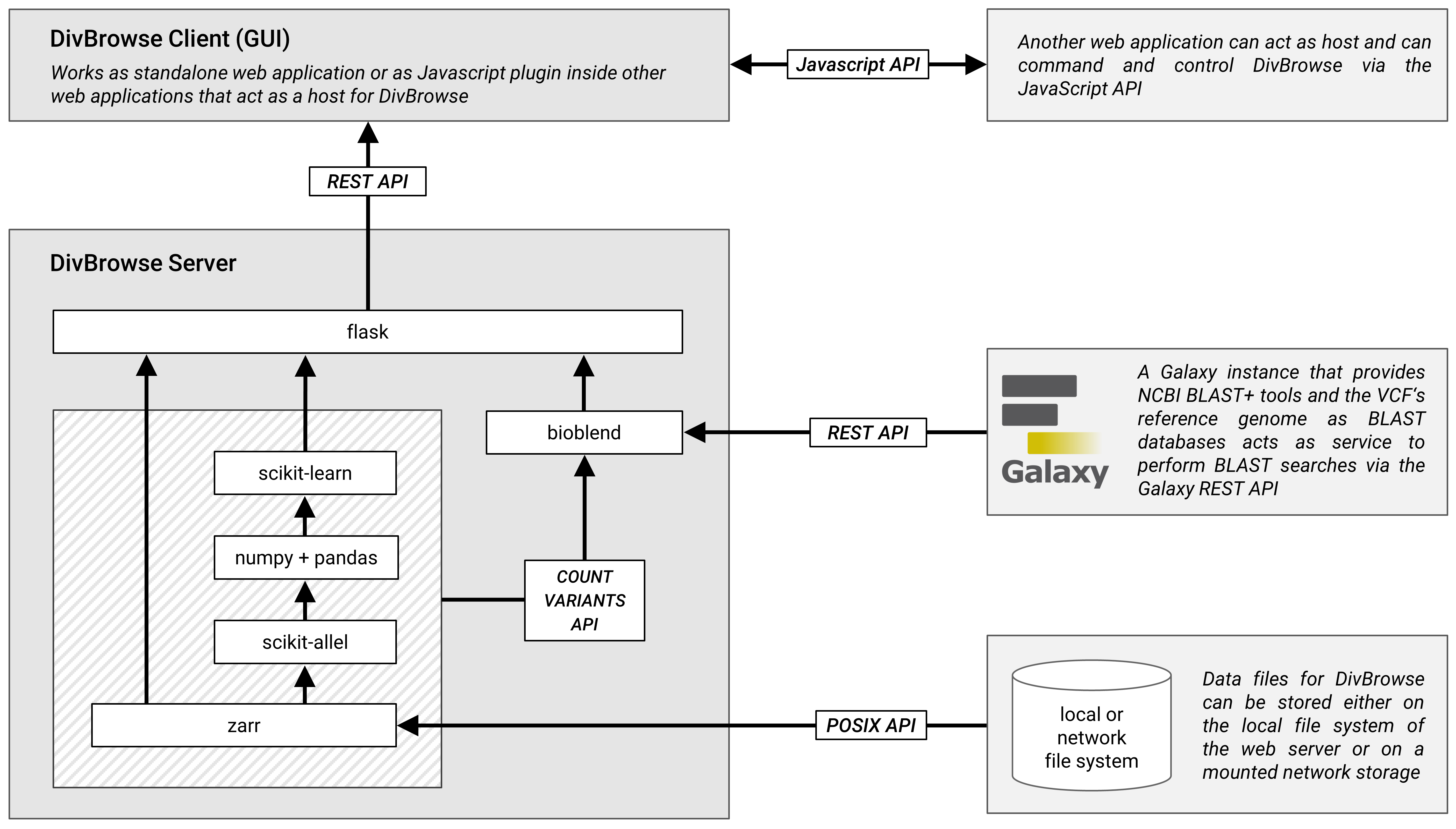
## Architecture
Raw data
{
"_id": null,
"home_page": "https://divbrowse.ipk-gatersleben.de/",
"name": "divbrowse",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.9,<4.0",
"maintainer_email": "",
"keywords": "bioinformatics,genomics,visualization,variant call format,vcf,biodiversity",
"author": "Patrick K\u00f6nig",
"author_email": "koenig@ipk-gatersleben.de",
"download_url": "https://files.pythonhosted.org/packages/92/d4/a6ecb55ef5b5e608387a7517c40296fdbb792e4780539d755ebd7b8fdcdb/divbrowse-1.1.0.tar.gz",
"platform": null,
"description": "<img src=\"docs/source/images/divbrowse_logo.png\" width=\"600\">\n<br />\n\n[](https://pypi.org/project/divbrowse/)\n[](https://hub.docker.com/r/ipkbit/divbrowse)\n\n\n[](https://doi.org/10.1093/gigascience/giad025)\n\n\n[](https://divbrowse.readthedocs.io/?badge=latest)\n[](https://badge.fury.io/py/divbrowse)\n[](https://pypi.org/project/divbrowse/)\n[](https://libraries.io/pypi/divbrowse)\n\n\n<br />\n\n**Website:** https://divbrowse.ipk-gatersleben.de \n**Documentation:** https://divbrowse.readthedocs.io\n**Paper:** https://doi.org/10.1093/gigascience/giad025\n\n<hr />\n\n**Table of contents:**\n- [About DivBrowse](#about-divbrowse)\n- [Try out DivBrowse](#try-out-divbrowse)\n- [Screenshots](#screenshots)\n- [Usage workflow concept](#usage-workflow-concept)\n- [Architecture](#architecture)\n\n<br />\n\n## About DivBrowse\n\nDivBrowse is a web application for interactive exploration and analysis of very large SNP matrices.\n\nIt offers a novel approach for interactive visualization and analysis of genomic diversity data and optionally also gene annotation data. The use of standard file formats for data input supports interoperability and seamless deployment of application instances based on established bioinformatics pipelines. The possible integration into 3rd-party web applications supports interoperability and reusability.\n\nThe integrated ad-hoc calculation of variant summary statistics and principal component analysis enables the user to perform interactive analysis of population structure for single genetic features like genes, exons and promoter regions. Data interoperability is achieved by the possibility to export genomic diversity data for genomic regions of interest in standardized VCF files.\n\n\n## Try out DivBrowse\n\nIf you want to test DivBrowse please visit the demo instances listed here:\nhttps://divbrowse.ipk-gatersleben.de/#demo-instances\n\n\n## Screenshots\n\n\n\n\n## Usage workflow concept\n\n\n\n\n## Architecture\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A web application for interactive visualization and analysis of genotypic variant matrices",
"version": "1.1.0",
"project_urls": {
"Documentation": "https://divbrowse.readthedocs.io/",
"Homepage": "https://divbrowse.ipk-gatersleben.de/",
"Repository": "https://github.com/IPK-BIT/divbrowse"
},
"split_keywords": [
"bioinformatics",
"genomics",
"visualization",
"variant call format",
"vcf",
"biodiversity"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "0ae73fd3db5a447011883cab9f42ec60ce6d3f78fa34df13ad1f9fd8927f90a6",
"md5": "1c723148b2cec8b4e3f36e8fd4f04df8",
"sha256": "c836d544d4bedaf2ba6a773eb8cd26acba628ddfd9a941f4a3df4349fba84d05"
},
"downloads": -1,
"filename": "divbrowse-1.1.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "1c723148b2cec8b4e3f36e8fd4f04df8",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.9,<4.0",
"size": 1604054,
"upload_time": "2023-05-16T11:22:34",
"upload_time_iso_8601": "2023-05-16T11:22:34.949452Z",
"url": "https://files.pythonhosted.org/packages/0a/e7/3fd3db5a447011883cab9f42ec60ce6d3f78fa34df13ad1f9fd8927f90a6/divbrowse-1.1.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "92d4a6ecb55ef5b5e608387a7517c40296fdbb792e4780539d755ebd7b8fdcdb",
"md5": "e15fecb1f0275ca25bf737242e1f9fde",
"sha256": "b586902383420fe076920e7cad3f7c7ee516da51e2a1f4afd48f713c20e0ef01"
},
"downloads": -1,
"filename": "divbrowse-1.1.0.tar.gz",
"has_sig": false,
"md5_digest": "e15fecb1f0275ca25bf737242e1f9fde",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.9,<4.0",
"size": 2883447,
"upload_time": "2023-05-16T11:22:39",
"upload_time_iso_8601": "2023-05-16T11:22:39.387108Z",
"url": "https://files.pythonhosted.org/packages/92/d4/a6ecb55ef5b5e608387a7517c40296fdbb792e4780539d755ebd7b8fdcdb/divbrowse-1.1.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-05-16 11:22:39",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "IPK-BIT",
"github_project": "divbrowse",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "divbrowse"
}
