# ASCA: ANOVA-Simultaneous Component Analysis in Python
<!-- TABLE OF CONTENTS -->
## Table of Contents
* [About the Project](#about-the-project)
* [Getting Started](#getting-started)
* [Simple Examples](#simple-examples)
* [Contributing](#contributing)
* [License](#license)
* [Contact](#contact)
* [References](#references)
<!-- ABOUT THE PROJECT -->
## About The Project
ASCA is a multivariate approach to the standard ANOVA, using simultaneous component analysis to interprete the underlying factors and interaction from a design of experiment dataset. This project implements ASCA in python to support open source development and a wider application of ASCA.
<!-- GETTING STARTED -->
## Getting Started
Install this library either from the official pypi or from this Github repository:
```
pip install ASCA
```
## Install most updated version from Github
In a environment terminal or CMD:
```bat
pip install git+https://github.com/tsyet12/ASCA
```
### Simple Example
```python
X = [[1.0000,0.6000],
[3.0000,0.4000],
[2.0000,0.7000],
[1.0000,0.8000],
[2.0000,0.0100],
[2.0000,0.8000],
[4.0000,1.0000],
[6.0000,2.0000],
[5.0000,0.9000],
[5.0000,1.0000],
[6.0000,2.0000],
[5.0000,0.7000]]
X=np.asarray(X)
F = [[1, 1],
[1, 1],
[1, 2],
[1, 2],
[1, 3],
[1, 3],
[2, 1],
[2, 1],
[2, 2],
[2, 2],
[2, 3],
[2, 3]]
F=np.asarray(F)
interactions = [[0, 1]]
ASCA=ASCA()
ASCA.fit(X,F,interactions)
ASCA.plot_factors()
ASCA.plot_interactions()
```
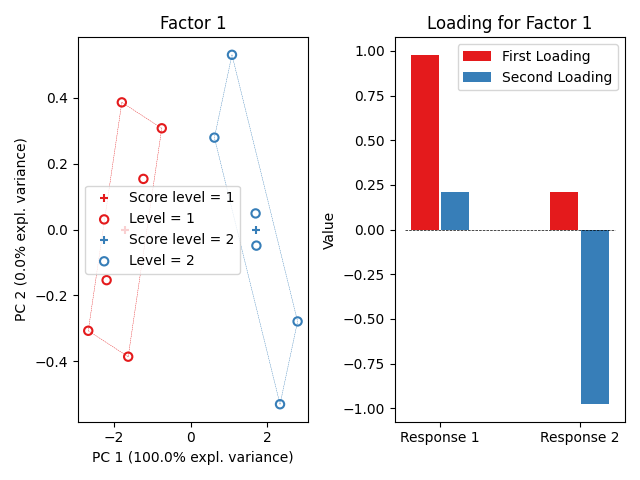
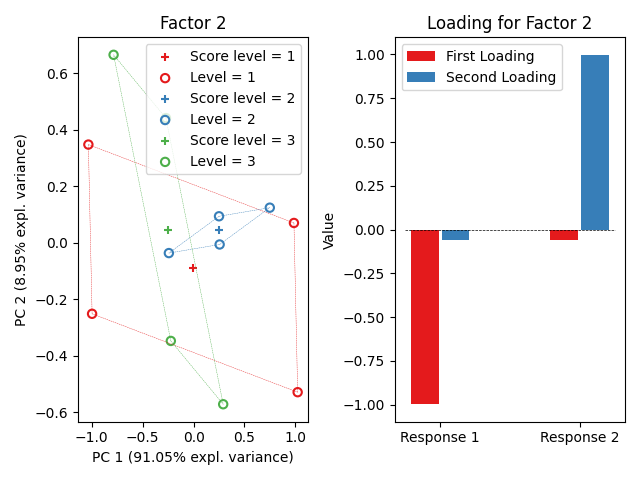
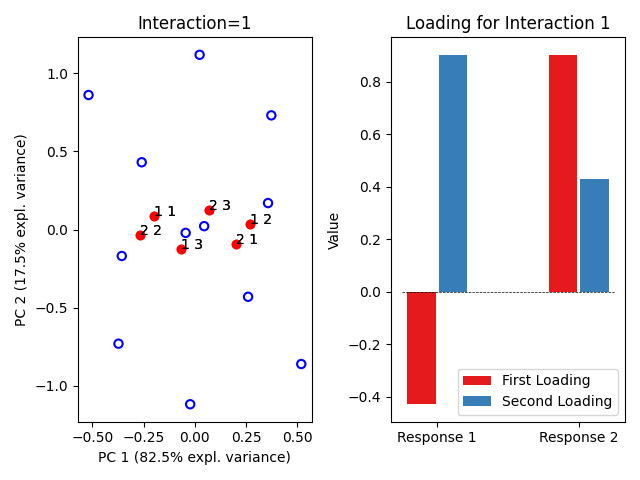
<!-- CONTRIBUTING -->
## Contributing
Contributions are what make the open source community such an amazing place to be learn, inspire, and create. Any contributions you make are **greatly appreciated**.
1. Fork the Project
2. Create your Feature Branch (`git checkout -b testbranch/prep`)
3. Commit your Changes (`git commit -m 'Improve testbranch/prep'`)
4. Push to the Branch (`git push origin testbranch/prep`)
5. Open a Pull Request
<!-- LICENSE -->
## License
Distributed under the Open Sourced BSD-2-Clause License. See [`LICENSE`](https://github.com/tsyet12/Chemsy/blob/main/LICENSE) for more information.
<!-- CONTACT -->
## Contact
Main Developer:
Sin Yong Teng sinyong.teng@ru.nl or tsyet12@gmail.com
Radboud University Nijmegen
<!-- References -->
## References
Smilde, Age K., et al. "ANOVA-simultaneous component analysis (ASCA): a new tool for analyzing designed metabolomics data." Bioinformatics 21.13 (2005): 3043-3048.
Jansen, Jeroen J., et al. "ASCA: analysis of multivariate data obtained from an experimental design." Journal of Chemometrics: A Journal of the Chemometrics Society 19.9 (2005): 469-481.
## Acknowledgements
The research contribution from S.Y. Teng is supported by the European Union's Horizon Europe Research and Innovation Program, under Marie Skłodowska-Curie Actions grant agreement no. 101064585 (MoCEGS).
Raw data
{
"_id": null,
"home_page": null,
"name": "ASCA",
"maintainer": null,
"docs_url": null,
"requires_python": null,
"maintainer_email": null,
"keywords": "Design of Experiments, Chemometrics, Artificial Intelligence",
"author": "Sin Yong Teng",
"author_email": "tsyet12@gmail.com",
"download_url": "https://files.pythonhosted.org/packages/86/3d/7098457b6d12adffb85d06ddee7b989453001b18aaa643dca46a5464b161/ASCA-1.1.tar.gz",
"platform": null,
"description": "# ASCA: ANOVA-Simultaneous Component Analysis in Python\r\n\r\n\r\n<!-- TABLE OF CONTENTS -->\r\n## Table of Contents\r\n\r\n* [About the Project](#about-the-project)\r\n* [Getting Started](#getting-started)\r\n* [Simple Examples](#simple-examples)\r\n* [Contributing](#contributing)\r\n* [License](#license)\r\n* [Contact](#contact)\r\n* [References](#references)\r\n\r\n\r\n<!-- ABOUT THE PROJECT -->\r\n## About The Project\r\nASCA is a multivariate approach to the standard ANOVA, using simultaneous component analysis to interprete the underlying factors and interaction from a design of experiment dataset. This project implements ASCA in python to support open source development and a wider application of ASCA.\r\n\r\n\r\n<!-- GETTING STARTED -->\r\n## Getting Started\r\n\r\nInstall this library either from the official pypi or from this Github repository:\r\n```\r\npip install ASCA\r\n```\r\n\r\n## Install most updated version from Github\r\n\r\nIn a environment terminal or CMD:\r\n```bat\r\npip install git+https://github.com/tsyet12/ASCA\r\n```\r\n\r\n\r\n\r\n\r\n### Simple Example\r\n```python\r\n\r\n X = [[1.0000,0.6000], \r\n [3.0000,0.4000],\r\n [2.0000,0.7000],\r\n [1.0000,0.8000],\r\n [2.0000,0.0100],\r\n [2.0000,0.8000],\r\n [4.0000,1.0000],\r\n [6.0000,2.0000],\r\n [5.0000,0.9000],\r\n [5.0000,1.0000],\r\n [6.0000,2.0000],\r\n [5.0000,0.7000]]\r\n X=np.asarray(X)\r\n\r\n F = [[1, 1],\r\n [1, 1],\r\n [1, 2],\r\n [1, 2],\r\n [1, 3],\r\n [1, 3],\r\n [2, 1],\r\n [2, 1],\r\n [2, 2],\r\n [2, 2],\r\n [2, 3],\r\n [2, 3]]\r\n F=np.asarray(F)\r\n interactions = [[0, 1]]\r\n\r\n ASCA=ASCA()\r\n ASCA.fit(X,F,interactions)\r\n ASCA.plot_factors()\r\n ASCA.plot_interactions()\r\n\r\n```\r\n\r\n\r\n\r\n\r\n\r\n\r\n\r\n<!-- CONTRIBUTING -->\r\n## Contributing\r\n\r\nContributions are what make the open source community such an amazing place to be learn, inspire, and create. Any contributions you make are **greatly appreciated**.\r\n\r\n1. Fork the Project\r\n2. Create your Feature Branch (`git checkout -b testbranch/prep`)\r\n3. Commit your Changes (`git commit -m 'Improve testbranch/prep'`)\r\n4. Push to the Branch (`git push origin testbranch/prep`)\r\n5. Open a Pull Request\r\n\r\n\r\n<!-- LICENSE -->\r\n## License\r\n\r\nDistributed under the Open Sourced BSD-2-Clause License. See [`LICENSE`](https://github.com/tsyet12/Chemsy/blob/main/LICENSE) for more information.\r\n\r\n\r\n<!-- CONTACT -->\r\n## Contact\r\nMain Developer:\r\n\r\nSin Yong Teng sinyong.teng@ru.nl or tsyet12@gmail.com\r\nRadboud University Nijmegen\r\n\r\n<!-- References -->\r\n## References\r\nSmilde, Age K., et al. \"ANOVA-simultaneous component analysis (ASCA): a new tool for analyzing designed metabolomics data.\" Bioinformatics 21.13 (2005): 3043-3048.\r\n\r\nJansen, Jeroen J., et al. \"ASCA: analysis of multivariate data obtained from an experimental design.\" Journal of Chemometrics: A Journal of the Chemometrics Society 19.9 (2005): 469-481.\r\n\r\n\r\n## Acknowledgements\r\nThe research contribution from S.Y. Teng is supported by the European Union's Horizon Europe Research and Innovation Program, under Marie Sk\u0142odowska-Curie Actions grant agreement no. 101064585 (MoCEGS).\r\n",
"bugtrack_url": null,
"license": "BSD 2-Clause",
"summary": "ASCA: ANOVA-simultaneous component analysis",
"version": "1.1",
"project_urls": null,
"split_keywords": [
"design of experiments",
" chemometrics",
" artificial intelligence"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "863d7098457b6d12adffb85d06ddee7b989453001b18aaa643dca46a5464b161",
"md5": "6a8a348adad269e1585b4792d8ba83b6",
"sha256": "bc24ac28760a4993445df7466e4c0bd737b3e4d8ee25b21182e8a0a75a517b6f"
},
"downloads": -1,
"filename": "ASCA-1.1.tar.gz",
"has_sig": false,
"md5_digest": "6a8a348adad269e1585b4792d8ba83b6",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 80959,
"upload_time": "2024-05-15T16:21:18",
"upload_time_iso_8601": "2024-05-15T16:21:18.610531Z",
"url": "https://files.pythonhosted.org/packages/86/3d/7098457b6d12adffb85d06ddee7b989453001b18aaa643dca46a5464b161/ASCA-1.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-05-15 16:21:18",
"github": false,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"lcname": "asca"
}