# Introduction of BMINet
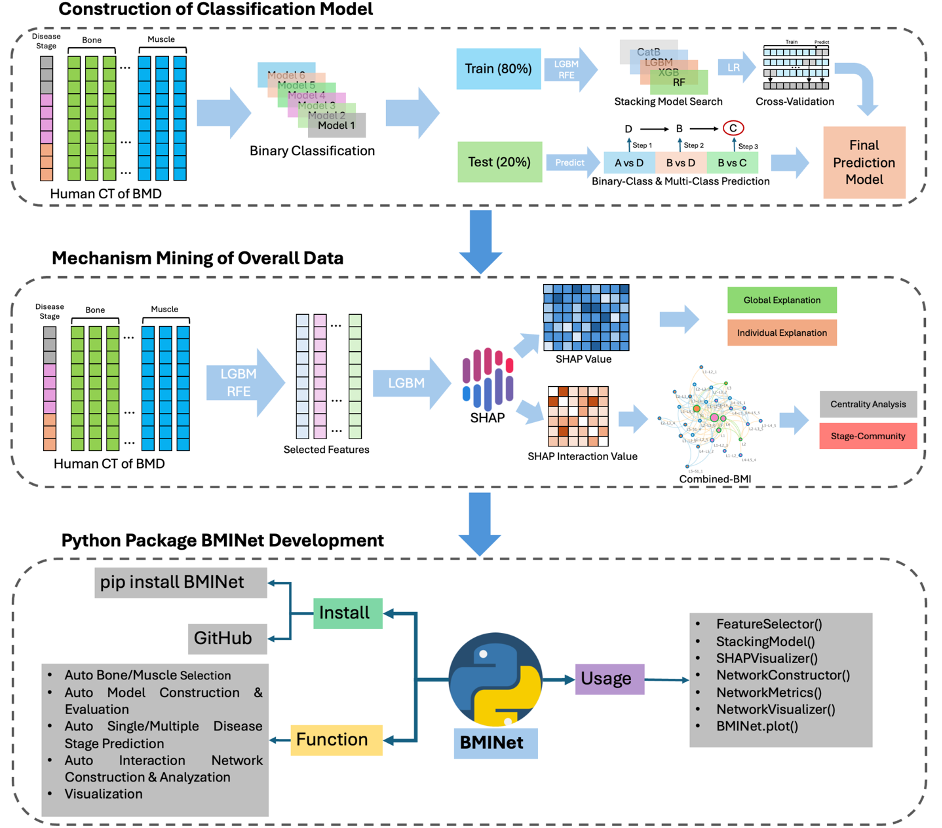
Machine learning and network based tool for:
- Classify and predict different disease stages precisely
- Understand machenism using SHAP model explanation
- Detect Bone-Muscle Interact network and detect disease modules
# Install
## Dependency
- **Python>=3.9**
## Quick install
- Install from pypi: `pip install BMINet`
- Or install from GitHub:
```
git clone https://github.com/Spencer-JRWang/BMINet
cd BMINet
pip install .
```
# Example Usage
- First, you should prepare your data (from CT or MRI etc.): [Example Data](https://github.com/Spencer-JRWang/BMINet/blob/main/Example/data.txt)
- Load your data
```python
import pandas as pd
df = pd.read_csv('Example/data.txt', sep='\t')
```
- Select features
```python
# Load FeatureSelector
from BMINet.Interaction import FeatureSelector
selector = FeatureSelector(core_name="LightGBM")
# Conduct feature selection from df
selected_features = selector.select(df)
```
> core_name should be "LightGBM", "XGBoost" or "CatBoost"
- Build linear formula model
```python
from BMINet.Model import Lasso_Formula
formula = Lasso_Formula(df, disease_pairs=("A", "B"))
print(formula)
```
> Also Ridge, ElasticNet and Simple Linear
- Build stacking model
```python
from BMINet.Interaction import StackingModel
# Load stacking model framework
'''
default: StackingModel(
base_models = [
('LGBM', LGBMClassifier(verbose=-1, n_estimators=1000, max_depth=5)),
('XGBoost', XGBClassifier(n_estimators=1000, max_depth=5)),
('CatBoost', CatBoostClassifier(verbose=False, iterations=800, max_depth=5))
]),
meta_model = LogisticRegression()
'''
Model = StackingModel()
best_scores = Model.stacking_model_search(df, selected_features)
```
> Using default base model and meta model, you can also define it yourself
- Predict of each group
```python
# single predict
single_prediction_score = Model.single_predict("A vs B", [...], use_our_model=True)
# multiple predict
multiple_prediction_score = Model.multiple_predict_predict("A vs B", [[...], [...],], use_our_model=True)
```
> If you use `use_our model = True`, you are predicting disease staged based on our data and model
>
> If you are researching on a brand new project, use `use_our_model = False`
- Basic machine learning plots
```python
from BMINet.plot import plot_ml_roc
plot_ml_roc(best_scores)
from BMINet.plot import plot_precision_recall
plot_precision_recall(best_scores)
from BMINet.plot import plot_score_histogram
plot_score_histogram(best_scores)
from BMINet.plot import plot_calibration_curve
plot_calibration_curve(best_scores)
```
- Model Explanation
```python
from BMINet.Interaction import SHAPVisualizer
# Load explanation class and train it
shap_visualizer = SHAPVisualizer(core_name="LightGBM")
shap_visualizer.train_model(df, selected_features)
# The dir you want to save the files
shap_visualizer.plot_shap('./Example')
shap_visualizer.plot_dependence('./Example')
```
- Network Construction
```python
# Load NetworkConstructor
from BMINet.Interaction import NetworkConstructor
network_constructor = NetworkConstructor(core_name="LightGBM", cutoff = 1.5)
# Construct sub-network list for each group
interactions = network_constructor.construct_network(df, selected_features)
# Construct conmbined network
combined_graph = network_constructor.compose_all(interactions)
# Remove isolated nodes from the network
Graph_BMI = network_constructor.remove_isolated_nodes(combined_graph)
# Save to .graphml file
network_constructor.save_graph(Graph_BMI, './Example')
```
- Network Analysis
```python
from BMINet.Interaction import NetworkMetrics
metrics_calculator = NetworkMetrics(Graph_BMI)
metrics = metrics_calculator.compute_metrics()
```
> You can see more Example usage at [here](https://github.com/Spencer-JRWang/BMINet/blob/main/Example.ipynb)
# Update Log
- 2024/9/5: Version `0.1.0`, fix CatBoost bug
- 2024/9/5: Version `0.1.1`, update prediction threholds
- 2024/11/8: Version `0.1.2`, update linear formula model
- 2024/11/26: Version `0.1.3`, add matrix visulization
> BMINet is developed by Jingran Wang
> Department of Orthopedic Surgery, The First Affiliated Hospital of Soochow University
> MOE Key Laboratory of Geriatric Diseases and Immunology, Suzhou Key Laboratory of Pathogen Bioscience and Anti-Infective Medicine
---
<div style="display: flex; justify-content: space-between; align-items: center;">
<img src="https://raw.githubusercontent.com/Spencer-JRWang/BMINet/main/Example/LOGO.png" alt="Organization Logo" style="width: 100px; height: auto; margin-left: 20px;">
</div>
Raw data
{
"_id": null,
"home_page": "https://github.com/Spencer-JRWang/BMINet",
"name": "BMINet",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.9",
"maintainer_email": null,
"keywords": "python, BMINet, Interaction, Network, Bone-Muscle, windows, mac, linux",
"author": "Spencer Wang",
"author_email": "jrwangspencer@stu.suda.edu.cn",
"download_url": "https://files.pythonhosted.org/packages/1a/2d/5c019a9c55f378e40f48204ff19bfd13ae5dd7c2654fe4802ad2537a896c/bminet-0.1.4.tar.gz",
"platform": null,
"description": "\n# Introduction of BMINet\n\nMachine learning and network based tool for:\n- Classify and predict different disease stages precisely\n- Understand machenism using SHAP model explanation\n- Detect Bone-Muscle Interact network and detect disease modules\n# Install\n## Dependency\n- **Python>=3.9**\n## Quick install\n- Install from pypi: `pip install BMINet`\n- Or install from GitHub: \n```\ngit clone https://github.com/Spencer-JRWang/BMINet\ncd BMINet\npip install .\n```\n# Example Usage\n- First, you should prepare your data (from CT or MRI etc.): [Example Data](https://github.com/Spencer-JRWang/BMINet/blob/main/Example/data.txt)\n- Load your data\n```python\nimport pandas as pd\ndf = pd.read_csv('Example/data.txt', sep='\\t')\n```\n- Select features\n```python\n# Load FeatureSelector\nfrom BMINet.Interaction import FeatureSelector\nselector = FeatureSelector(core_name=\"LightGBM\")\n# Conduct feature selection from df\nselected_features = selector.select(df)\n```\n> core_name should be \"LightGBM\", \"XGBoost\" or \"CatBoost\"\n\n- Build linear formula model\n```python\nfrom BMINet.Model import Lasso_Formula\nformula = Lasso_Formula(df, disease_pairs=(\"A\", \"B\"))\nprint(formula)\n```\n> Also Ridge, ElasticNet and Simple Linear\n\n- Build stacking model\n```python\nfrom BMINet.Interaction import StackingModel\n# Load stacking model framework\n'''\ndefault: StackingModel(\n base_models = [\n ('LGBM', LGBMClassifier(verbose=-1, n_estimators=1000, max_depth=5)),\n ('XGBoost', XGBClassifier(n_estimators=1000, max_depth=5)),\n ('CatBoost', CatBoostClassifier(verbose=False, iterations=800, max_depth=5))\n ]), \n meta_model = LogisticRegression()\n'''\nModel = StackingModel()\nbest_scores = Model.stacking_model_search(df, selected_features)\n```\n> Using default base model and meta model, you can also define it yourself\n\n- Predict of each group\n```python\n# single predict\nsingle_prediction_score = Model.single_predict(\"A vs B\", [...], use_our_model=True)\n# multiple predict\nmultiple_prediction_score = Model.multiple_predict_predict(\"A vs B\", [[...], [...],], use_our_model=True)\n```\n> If you use `use_our model = True`, you are predicting disease staged based on our data and model\n> \n> If you are researching on a brand new project, use `use_our_model = False`\n\n- Basic machine learning plots\n```python\nfrom BMINet.plot import plot_ml_roc\nplot_ml_roc(best_scores)\nfrom BMINet.plot import plot_precision_recall\nplot_precision_recall(best_scores)\nfrom BMINet.plot import plot_score_histogram\nplot_score_histogram(best_scores)\nfrom BMINet.plot import plot_calibration_curve\nplot_calibration_curve(best_scores)\n```\n- Model Explanation\n```python\nfrom BMINet.Interaction import SHAPVisualizer\n# Load explanation class and train it\nshap_visualizer = SHAPVisualizer(core_name=\"LightGBM\")\nshap_visualizer.train_model(df, selected_features)\n# The dir you want to save the files\nshap_visualizer.plot_shap('./Example')\nshap_visualizer.plot_dependence('./Example')\n```\n\n- Network Construction\n```python\n# Load NetworkConstructor\nfrom BMINet.Interaction import NetworkConstructor\nnetwork_constructor = NetworkConstructor(core_name=\"LightGBM\", cutoff = 1.5)\n\n# Construct sub-network list for each group\ninteractions = network_constructor.construct_network(df, selected_features)\n# Construct conmbined network\ncombined_graph = network_constructor.compose_all(interactions)\n# Remove isolated nodes from the network\nGraph_BMI = network_constructor.remove_isolated_nodes(combined_graph)\n# Save to .graphml file\nnetwork_constructor.save_graph(Graph_BMI, './Example')\n```\n\n- Network Analysis\n```python\nfrom BMINet.Interaction import NetworkMetrics\nmetrics_calculator = NetworkMetrics(Graph_BMI)\nmetrics = metrics_calculator.compute_metrics()\n```\n\n> You can see more Example usage at [here](https://github.com/Spencer-JRWang/BMINet/blob/main/Example.ipynb)\n\n# Update Log\n- 2024/9/5: Version `0.1.0`, fix CatBoost bug\n- 2024/9/5: Version `0.1.1`, update prediction threholds\n- 2024/11/8: Version `0.1.2`, update linear formula model\n- 2024/11/26: Version `0.1.3`, add matrix visulization\n\n> BMINet is developed by Jingran Wang\n\n> Department of Orthopedic Surgery, The First Affiliated Hospital of Soochow University\n\n> MOE Key Laboratory of Geriatric Diseases and Immunology, Suzhou Key Laboratory of Pathogen Bioscience and Anti-Infective Medicine\n---\n<div style=\"display: flex; justify-content: space-between; align-items: center;\">\n <img src=\"https://raw.githubusercontent.com/Spencer-JRWang/BMINet/main/Example/LOGO.png\" alt=\"Organization Logo\" style=\"width: 100px; height: auto; margin-left: 20px;\">\n</div>\n",
"bugtrack_url": null,
"license": null,
"summary": "Machine Learning and Graph based tool for detecting and analyzing Bone-Muscle Interactions",
"version": "0.1.4",
"project_urls": {
"Homepage": "https://github.com/Spencer-JRWang/BMINet"
},
"split_keywords": [
"python",
" bminet",
" interaction",
" network",
" bone-muscle",
" windows",
" mac",
" linux"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "1a2d5c019a9c55f378e40f48204ff19bfd13ae5dd7c2654fe4802ad2537a896c",
"md5": "5b774ae46561c8a49f8035f5e7afec0a",
"sha256": "b8856a9ae0b46ee78c9b8cc436c699eff2e70e3a3abae3fd178c55c1c0d92c5f"
},
"downloads": -1,
"filename": "bminet-0.1.4.tar.gz",
"has_sig": false,
"md5_digest": "5b774ae46561c8a49f8035f5e7afec0a",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.9",
"size": 118521,
"upload_time": "2024-12-15T06:35:28",
"upload_time_iso_8601": "2024-12-15T06:35:28.405886Z",
"url": "https://files.pythonhosted.org/packages/1a/2d/5c019a9c55f378e40f48204ff19bfd13ae5dd7c2654fe4802ad2537a896c/bminet-0.1.4.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-12-15 06:35:28",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "Spencer-JRWang",
"github_project": "BMINet",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"requirements": [
{
"name": "anyio",
"specs": [
[
"==",
"4.4.0"
]
]
},
{
"name": "argon2-cffi",
"specs": [
[
"==",
"23.1.0"
]
]
},
{
"name": "argon2-cffi-bindings",
"specs": [
[
"==",
"21.2.0"
]
]
},
{
"name": "arrow",
"specs": [
[
"==",
"1.3.0"
]
]
},
{
"name": "async-lru",
"specs": [
[
"==",
"2.0.4"
]
]
},
{
"name": "attrs",
"specs": [
[
"==",
"24.2.0"
]
]
},
{
"name": "babel",
"specs": [
[
"==",
"2.16.0"
]
]
},
{
"name": "beautifulsoup4",
"specs": [
[
"==",
"4.12.3"
]
]
},
{
"name": "bleach",
"specs": [
[
"==",
"6.1.0"
]
]
},
{
"name": "blinker",
"specs": [
[
"==",
"1.8.2"
]
]
},
{
"name": "catboost",
"specs": [
[
"==",
"1.2.5"
]
]
},
{
"name": "certifi",
"specs": [
[
"==",
"2024.8.30"
]
]
},
{
"name": "cffi",
"specs": [
[
"==",
"1.17.1"
]
]
},
{
"name": "charset-normalizer",
"specs": [
[
"==",
"3.3.2"
]
]
},
{
"name": "click",
"specs": [
[
"==",
"8.1.7"
]
]
},
{
"name": "cloudpickle",
"specs": [
[
"==",
"3.0.0"
]
]
},
{
"name": "community",
"specs": [
[
"==",
"1.0.0b1"
]
]
},
{
"name": "contourpy",
"specs": [
[
"==",
"1.3.0"
]
]
},
{
"name": "cycler",
"specs": [
[
"==",
"0.12.1"
]
]
},
{
"name": "defusedxml",
"specs": [
[
"==",
"0.7.1"
]
]
},
{
"name": "fastjsonschema",
"specs": [
[
"==",
"2.20.0"
]
]
},
{
"name": "Flask",
"specs": [
[
"==",
"3.0.3"
]
]
},
{
"name": "fonttools",
"specs": [
[
"==",
"4.53.1"
]
]
},
{
"name": "fqdn",
"specs": [
[
"==",
"1.5.1"
]
]
},
{
"name": "graphviz",
"specs": [
[
"==",
"0.20.3"
]
]
},
{
"name": "h11",
"specs": [
[
"==",
"0.14.0"
]
]
},
{
"name": "httpcore",
"specs": [
[
"==",
"1.0.5"
]
]
},
{
"name": "httpx",
"specs": [
[
"==",
"0.27.2"
]
]
},
{
"name": "idna",
"specs": [
[
"==",
"3.8"
]
]
},
{
"name": "ipywidgets",
"specs": [
[
"==",
"8.1.5"
]
]
},
{
"name": "isoduration",
"specs": [
[
"==",
"20.11.0"
]
]
},
{
"name": "itsdangerous",
"specs": [
[
"==",
"2.2.0"
]
]
},
{
"name": "Jinja2",
"specs": [
[
"==",
"3.1.4"
]
]
},
{
"name": "joblib",
"specs": [
[
"==",
"1.4.2"
]
]
},
{
"name": "json5",
"specs": [
[
"==",
"0.9.25"
]
]
},
{
"name": "jsonpointer",
"specs": [
[
"==",
"3.0.0"
]
]
},
{
"name": "jsonschema",
"specs": [
[
"==",
"4.23.0"
]
]
},
{
"name": "jsonschema-specifications",
"specs": [
[
"==",
"2023.12.1"
]
]
},
{
"name": "jupyter",
"specs": [
[
"==",
"1.1.1"
]
]
},
{
"name": "jupyter-console",
"specs": [
[
"==",
"6.6.3"
]
]
},
{
"name": "jupyter-events",
"specs": [
[
"==",
"0.10.0"
]
]
},
{
"name": "jupyter-lsp",
"specs": [
[
"==",
"2.2.5"
]
]
},
{
"name": "jupyter_server",
"specs": [
[
"==",
"2.14.2"
]
]
},
{
"name": "jupyter_server_terminals",
"specs": [
[
"==",
"0.5.3"
]
]
},
{
"name": "jupyterlab",
"specs": [
[
"==",
"4.2.5"
]
]
},
{
"name": "jupyterlab_pygments",
"specs": [
[
"==",
"0.3.0"
]
]
},
{
"name": "jupyterlab_server",
"specs": [
[
"==",
"2.27.3"
]
]
},
{
"name": "jupyterlab_widgets",
"specs": [
[
"==",
"3.0.13"
]
]
},
{
"name": "kiwisolver",
"specs": [
[
"==",
"1.4.5"
]
]
},
{
"name": "lightgbm",
"specs": [
[
"==",
"4.5.0"
]
]
},
{
"name": "llvmlite",
"specs": [
[
"==",
"0.43.0"
]
]
},
{
"name": "MarkupSafe",
"specs": [
[
"==",
"2.1.5"
]
]
},
{
"name": "matplotlib",
"specs": [
[
"==",
"3.9.2"
]
]
},
{
"name": "mistune",
"specs": [
[
"==",
"3.0.2"
]
]
},
{
"name": "nbclient",
"specs": [
[
"==",
"0.10.0"
]
]
},
{
"name": "nbconvert",
"specs": [
[
"==",
"7.16.4"
]
]
},
{
"name": "nbformat",
"specs": [
[
"==",
"5.10.4"
]
]
},
{
"name": "networkx",
"specs": [
[
"==",
"3.3"
]
]
},
{
"name": "notebook",
"specs": [
[
"==",
"7.2.2"
]
]
},
{
"name": "notebook_shim",
"specs": [
[
"==",
"0.2.4"
]
]
},
{
"name": "numba",
"specs": [
[
"==",
"0.60.0"
]
]
},
{
"name": "numpy",
"specs": [
[
"==",
"1.26.0"
]
]
},
{
"name": "overrides",
"specs": [
[
"==",
"7.7.0"
]
]
},
{
"name": "pandas",
"specs": [
[
"==",
"2.2.2"
]
]
},
{
"name": "pandocfilters",
"specs": [
[
"==",
"1.5.1"
]
]
},
{
"name": "pillow",
"specs": [
[
"==",
"10.4.0"
]
]
},
{
"name": "plotly",
"specs": [
[
"==",
"5.24.0"
]
]
},
{
"name": "prometheus_client",
"specs": [
[
"==",
"0.20.0"
]
]
},
{
"name": "pycparser",
"specs": [
[
"==",
"2.22"
]
]
},
{
"name": "pyparsing",
"specs": [
[
"==",
"3.1.4"
]
]
},
{
"name": "python-json-logger",
"specs": [
[
"==",
"2.0.7"
]
]
},
{
"name": "pytz",
"specs": [
[
"==",
"2024.1"
]
]
},
{
"name": "PyYAML",
"specs": [
[
"==",
"6.0.2"
]
]
},
{
"name": "referencing",
"specs": [
[
"==",
"0.35.1"
]
]
},
{
"name": "requests",
"specs": [
[
"==",
"2.32.3"
]
]
},
{
"name": "rfc3339-validator",
"specs": [
[
"==",
"0.1.4"
]
]
},
{
"name": "rfc3986-validator",
"specs": [
[
"==",
"0.1.1"
]
]
},
{
"name": "rpds-py",
"specs": [
[
"==",
"0.20.0"
]
]
},
{
"name": "scikit-learn",
"specs": [
[
"==",
"1.4.0"
]
]
},
{
"name": "scipy",
"specs": [
[
"==",
"1.14.1"
]
]
},
{
"name": "seaborn",
"specs": [
[
"==",
"0.13.2"
]
]
},
{
"name": "Send2Trash",
"specs": [
[
"==",
"1.8.3"
]
]
},
{
"name": "setuptools",
"specs": [
[
"==",
"73.0.1"
]
]
},
{
"name": "shap",
"specs": [
[
"==",
"0.46.0"
]
]
},
{
"name": "slicer",
"specs": [
[
"==",
"0.0.8"
]
]
},
{
"name": "sniffio",
"specs": [
[
"==",
"1.3.1"
]
]
},
{
"name": "soupsieve",
"specs": [
[
"==",
"2.6"
]
]
},
{
"name": "tenacity",
"specs": [
[
"==",
"9.0.0"
]
]
},
{
"name": "terminado",
"specs": [
[
"==",
"0.18.1"
]
]
},
{
"name": "threadpoolctl",
"specs": [
[
"==",
"3.5.0"
]
]
},
{
"name": "tinycss2",
"specs": [
[
"==",
"1.3.0"
]
]
},
{
"name": "tqdm",
"specs": [
[
"==",
"4.66.5"
]
]
},
{
"name": "types-python-dateutil",
"specs": [
[
"==",
"2.9.0.20240906"
]
]
},
{
"name": "tzdata",
"specs": [
[
"==",
"2024.1"
]
]
},
{
"name": "uri-template",
"specs": [
[
"==",
"1.3.0"
]
]
},
{
"name": "urllib3",
"specs": [
[
"==",
"2.2.2"
]
]
},
{
"name": "webcolors",
"specs": [
[
"==",
"24.8.0"
]
]
},
{
"name": "webencodings",
"specs": [
[
"==",
"0.5.1"
]
]
},
{
"name": "websocket-client",
"specs": [
[
"==",
"1.8.0"
]
]
},
{
"name": "Werkzeug",
"specs": [
[
"==",
"3.0.4"
]
]
},
{
"name": "wheel",
"specs": [
[
"==",
"0.44.0"
]
]
},
{
"name": "widgetsnbextension",
"specs": [
[
"==",
"4.0.13"
]
]
},
{
"name": "xgboost",
"specs": [
[
"==",
"2.1.1"
]
]
}
],
"lcname": "bminet"
}
