[](https://brats-toolkit.readthedocs.io/en/latest/?badge=latest)
[](https://pypi.python.org/pypi/BraTS-Toolkit/)
____ _____ ____ _____ _ _ _ _
| __ ) _ __ __ |_ _/ ___| |_ _|__ ___ | | | _(_) |_
| _ \| '__/ _` || | \___ \ | |/ _ \ / _ \| | |/ / | __|
| |_) | | | (_| || | ___) | | | (_) | (_) | | <| | |_
|____/|_| \__,_||_| |____/ |_|\___/ \___/|_|_|\_\_|\__|
# BraTS Toolkit: What is it and what can I use it for?
Abstract:
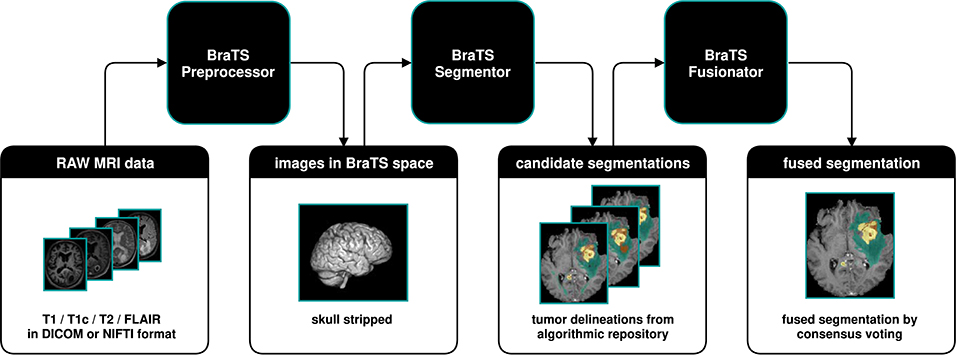
>BraTS Toolkit is a holistic approach to brain tumor segmentation and consists out of out of three components:
First, the BraTS Preprocessor facilitates data standardization and preprocessing for researchers and clinicians alike. It covers the entire image analysis workflow prior to tumor segmentation, from image conversion and registration to brain extraction. Second, BraTS Segmentor enables orchestration of BraTS brain tumor segmentation algorithms for generation of fully-automated segmentations. Finally, BraTS Fusionator can combine the resulting candidate segmentations into consensus segmentations using fusion methods such as majority voting and iterative SIMPLE fusion. The capabilities of our tools are illustrated with a practical example to enable easy translation to clinical and scientific practice.
## Installation
To install the most up-to-date version from the master branch, please use the following pip install command:
```
pip install BraTS-Toolkit
```
We recommended installation in a virtual environment based on Python 3.10 .
Further, NVIDIA Docker Toolkit needs to be installed (installation instructions here: https://docs.nvidia.com/datacenter/cloud-native/container-toolkit/install-guide.html#docker and here: https://neuronflow.github.io/BraTS-Preprocessor/#dockerinstallation ).
## Issues
When running into issues, please use the issue tracker here on Github: https://github.com/neuronflow/BraTS-Toolkit/issues
So others can profit and contribute as well.
## Citation
If you use BraTS Toolkit, please cite:
https://www.frontiersin.org/articles/10.3389/fnins.2020.00125/full
Kofler, F., Berger, C., Waldmannstetter, D., Lipkova, J., Ezhov, I., Tetteh, G., Kirschke, J., Zimmer, C., Wiestler, B., & Menze, B. H. (2020). BraTS Toolkit: Translating BraTS Brain Tumor Segmentation Algorithms Into Clinical and Scientific Practice. Frontiers in neuroscience, 14, 125. https://doi.org/10.3389/fnins.2020.00125
```
@article{kofler2020brats,
title={BraTS toolkit: translating BraTS brain tumor segmentation algorithms into clinical and scientific practice},
author={Kofler, Florian and Berger, Christoph and Waldmannstetter, Diana and Lipkova, Jana and Ezhov, Ivan and Tetteh, Giles and Kirschke, Jan and Zimmer, Claus and Wiestler, Benedikt and Menze, Bjoern H},
journal={Frontiers in neuroscience},
pages={125},
year={2020},
publisher={Frontiers}
}
```
Please also cite the following original authors of the algorithms who make this repository and tool possible:
| Docker Image | Paper |
| ---------------------- | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| econib | Marcinkiewicz, M., Nalepa, J., Lorenzo, P., Dudzik, W., & Mrukwa, G. (2018). Segmenting brain tumors from MRI using cascaded multi-modal U-Nets. In International MICCAI Brainlesion Workshop (pp. 13–24). |
| mic-dkfz | Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., & Maier-Hein, K. (2018). No new-net. In International MICCAI Brainlesion Workshop (pp. 234–244). |
| scan | McKinley, R., Meier, R., & Wiest, R. (2018). Ensembles of densely-connected CNNs with label-uncertainty for brain tumor segmentation. In International MICCAI Brainlesion Workshop (pp. 456–465). |
| xfeng | Feng, X., Tustison, N., & Meyer, C. (2019). Brain Tumor Segmentation Using an Ensemble of 3D U-Nets and Overall Survival Prediction Using Radiomic Features. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 279–288). Springer International Publishing. |
| lfb_rwth | Weninger, L., Rippel, O., Koppers, S., & Merhof, D. (2019). Segmentation of Brain Tumors and Patient Survival Prediction: Methods for the BraTS 2018 Challenge. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 3–12). Springer International Publishing. |
| gbmnet | Nuechterlein, N., & Mehta, S. (2019). 3D-ESPNet with Pyramidal Refinement for Volumetric Brain Tumor Image Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 245–253). Springer International Publishing. |
| zyx_2019 | Zhao, Y.X., Zhang, Y.M., Song, M., & Liu, C.L. (2019). Multi-view Semi-supervised 3D Whole Brain Segmentation with a Self-ensemble Network. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2019 (pp. 256–265). Springer International Publishing. |
| scan_2019 | McKinley, R., Rebsamen, M., Meier, R., & Wiest, R. (2020). Triplanar Ensemble of 3D-to-2D CNNs with Label-Uncertainty for Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 379–387). Springer International Publishing. |
| isen-20 | Isensee, F., Jäger, P. F., Full, P. M., Vollmuth, P., & Maier-Hein, K. H. (2021). nnU-Net for brain tumor segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, (pp. 118-132). Springer International Publishing. |
| hnfnetv1-20 | Jia, H., Cai, W., Huang, H., & Xia, Y. (2021). H^ 2 2 NF-Net for Brain Tumor Segmentation Using Multimodal MR Imaging: 2nd Place Solution to BraTS Challenge 2020 Segmentation Task. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, (pp. 58-68). Springer International Publishing. |
| yixinmpl-20 | Wang, Y., Zhang, Y., Hou, F., Liu, Y., Tian, J., Zhong, C., ... & He, Z. (2021). Modality-pairing learning for brain tumor segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020,(pp. 230-240). Springer International Publishing. |
| sanet0-20 | Yuan, Y. (2021). Automatic brain tumor segmentation with scale attention network. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries BrainLes 2020, (pp. 285-294). Springer International Publishing. |
| scan-20 & scan_lite-20 | McKinley, R., Rebsamen, M., Dätwyler, K., Meier, R., Radojewski, P., & Wiest, R. (2021). Uncertainty-driven refinement of tumor-core segmentation using 3d-to-2d networks with label uncertainty. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, (pp. 401-411). Springer International Publishing. |
## Usage
BrainLes features [Jupyter notebook tutorials](https://github.com/BrainLesion/tutorials/tree/main/BraTS-Toolkit) for glioma segmentation with BraTS Toolkit.
Further, have a look at the Python examples in this repository for preprocessing (`0_preprocessing_single.py`), segmentation (`1_segmentation.py`) and fusion (`2_fusion.py`).
## Brats Segmentor
BraTS Segmentor enables orchestration of BraTS brain tumor segmentation algorithms for generation of fully-automated segmentations.
For segmentation, your files should be `preprocessed`, meaning they should be co-registered and skullstripped in SRI-24 space.
You can preprocess your files using [BrainLes preprocessing](https://github.com/BrainLesion/preprocessing) (recommended) or the deprecated preprocessor of BraTS Toolkit (see below).
### Python package
Please have a look at `1_segmentation.py` in this repository for a demo application.
### Command Line Interface (CLI)
Type `brats-segment -h` after installing the Python package to see available options.
## Brats Fusionator
BraTS Fusionator can combine the resulting candidate segmentations into consensus segmentations using fusion methods such as majority voting and iterative SIMPLE fusion.
### Python package
Please have a look at `2_fusion.py` in this repository for a demo application.
## Brats Preprocessor (deprecated)
BraTS Preprocessor facilitates data standardization and preprocessing for researchers and clinicians alike. It covers the entire image analysis workflow prior to tumor segmentation, from image conversion and registration to brain extraction.
> WARNING: BraTS Preprocessor is deprecated. It still works, but we recommended using [BrainLes preprocessing](https://github.com/BrainLesion/preprocessing) instead, which offers much more flexibility.
### Processing Modi
BraTS Preprocessor offers the following preprocessing modes:
#### GPU brain extraction: "gpu"
Activates HD-BET in CUDA mode. Only supported when using NVIDIA docker (Linux operating systems only for the moment).
#### CPU brain extraction: "cpu"
Activates HD-BET or ROBEX in CPU mode depending on available RAM.
#### ROBEX brain extraction: "robex"
Activates Robex brain extraction.
#### GPU defacing: "gpu_defacing"
Activates GPU defacing - not implemented yet falls back to `cpu_defacing` for the moment.
#### CPU defacing: "cpu_defacing"
Activates mri-deface to deface using CPU.
### Single vs. batch processing
BraTS preprocessor offers processing files in `batch` or `single` mode. Please have a look at the respective example scripts / CLI commands for information how to use them.
For faster computation, we strictly recommend the `batch` processing mode, which avoids the additional overhead of spawning and shutting down multiple docker containers and instead does all the processing in one container.
### Python package
Please have a look at `0_preprocessing_batch.py` and `0_preprocessing_single.py` in this repository for a demo application. You can download the example data by cloning this repository.
### Command Line Interface (CLI)
#### single file processing
Type `brats-preprocess -h` after installing the Python package to see available options.
#### batch file processing
Type `brats-batch-preprocess -h` after installing the Python package to see available options.
### Graphical User Interface (GUI)
You can find instructions to use and download the GUI variant here: https://neuronflow.github.io/BraTS-Preprocessor/
> WARNING: The GUI is not nicely maintained. We encourage you to use the Python package or the CLI instead.
### Command Line Interface (CLI)
Type `brats-fuse -h` after installing the Python package to see available options.
## Contact / Feedback / Questions
Open an issue in this git repository or contact us via email.
Raw data
{
"_id": null,
"home_page": "https://github.com/neuronflow/BraTS-Toolkit",
"name": "BraTS-Toolkit",
"maintainer": "Florian Kofler",
"docs_url": null,
"requires_python": "<4.0,>=3.10",
"maintainer_email": "florian.kofler@tum.de",
"keywords": "brain tumor, glioma, BraTS, segmentation, fusion, skullstripping, brain extraction",
"author": "Florian Kofler",
"author_email": "florian.kofler@tum.de",
"download_url": null,
"platform": null,
"description": "[](https://brats-toolkit.readthedocs.io/en/latest/?badge=latest)\n[](https://pypi.python.org/pypi/BraTS-Toolkit/)\n\n ____ _____ ____ _____ _ _ _ _ \n | __ ) _ __ __ |_ _/ ___| |_ _|__ ___ | | | _(_) |_ \n | _ \\| '__/ _` || | \\___ \\ | |/ _ \\ / _ \\| | |/ / | __|\n | |_) | | | (_| || | ___) | | | (_) | (_) | | <| | |_ \n |____/|_| \\__,_||_| |____/ |_|\\___/ \\___/|_|_|\\_\\_|\\__|\n \n# BraTS Toolkit: What is it and what can I use it for?\nAbstract:\n>BraTS Toolkit is a holistic approach to brain tumor segmentation and consists out of out of three components:\n First, the BraTS Preprocessor facilitates data standardization and preprocessing for researchers and clinicians alike. It covers the entire image analysis workflow prior to tumor segmentation, from image conversion and registration to brain extraction. Second, BraTS Segmentor enables orchestration of BraTS brain tumor segmentation algorithms for generation of fully-automated segmentations. Finally, BraTS Fusionator can combine the resulting candidate segmentations into consensus segmentations using fusion methods such as majority voting and iterative SIMPLE fusion. The capabilities of our tools are illustrated with a practical example to enable easy translation to clinical and scientific practice.\n\n\n\n\n## Installation\nTo install the most up-to-date version from the master branch, please use the following pip install command:\n```\npip install BraTS-Toolkit\n```\n\nWe recommended installation in a virtual environment based on Python 3.10 .\n\nFurther, NVIDIA Docker Toolkit needs to be installed (installation instructions here: https://docs.nvidia.com/datacenter/cloud-native/container-toolkit/install-guide.html#docker and here: https://neuronflow.github.io/BraTS-Preprocessor/#dockerinstallation ).\n\n## Issues\nWhen running into issues, please use the issue tracker here on Github: https://github.com/neuronflow/BraTS-Toolkit/issues\nSo others can profit and contribute as well.\n\n## Citation\nIf you use BraTS Toolkit, please cite:\n\nhttps://www.frontiersin.org/articles/10.3389/fnins.2020.00125/full\n\nKofler, F., Berger, C., Waldmannstetter, D., Lipkova, J., Ezhov, I., Tetteh, G., Kirschke, J., Zimmer, C., Wiestler, B., & Menze, B. H. (2020). BraTS Toolkit: Translating BraTS Brain Tumor Segmentation Algorithms Into Clinical and Scientific Practice. Frontiers in neuroscience, 14, 125. https://doi.org/10.3389/fnins.2020.00125\n\n```\n@article{kofler2020brats,\n title={BraTS toolkit: translating BraTS brain tumor segmentation algorithms into clinical and scientific practice},\n author={Kofler, Florian and Berger, Christoph and Waldmannstetter, Diana and Lipkova, Jana and Ezhov, Ivan and Tetteh, Giles and Kirschke, Jan and Zimmer, Claus and Wiestler, Benedikt and Menze, Bjoern H},\n journal={Frontiers in neuroscience},\n pages={125},\n year={2020},\n publisher={Frontiers}\n}\n```\n\nPlease also cite the following original authors of the algorithms who make this repository and tool possible:\n\n| Docker Image | Paper |\n| ---------------------- | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |\n| econib | Marcinkiewicz, M., Nalepa, J., Lorenzo, P., Dudzik, W., & Mrukwa, G. (2018). Segmenting brain tumors from MRI using cascaded multi-modal U-Nets. In International MICCAI Brainlesion Workshop (pp. 13\u201324). |\n| mic-dkfz | Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., & Maier-Hein, K. (2018). No new-net. In International MICCAI Brainlesion Workshop (pp. 234\u2013244). |\n| scan | McKinley, R., Meier, R., & Wiest, R. (2018). Ensembles of densely-connected CNNs with label-uncertainty for brain tumor segmentation. In International MICCAI Brainlesion Workshop (pp. 456\u2013465). |\n| xfeng | Feng, X., Tustison, N., & Meyer, C. (2019). Brain Tumor Segmentation Using an Ensemble of 3D U-Nets and Overall Survival Prediction Using Radiomic Features. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 279\u2013288). Springer International Publishing. |\n| lfb_rwth | Weninger, L., Rippel, O., Koppers, S., & Merhof, D. (2019). Segmentation of Brain Tumors and Patient Survival Prediction: Methods for the BraTS 2018 Challenge. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 3\u201312). Springer International Publishing. |\n| gbmnet | Nuechterlein, N., & Mehta, S. (2019). 3D-ESPNet with Pyramidal Refinement for Volumetric Brain Tumor Image Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 245\u2013253). Springer International Publishing. |\n| zyx_2019 | Zhao, Y.X., Zhang, Y.M., Song, M., & Liu, C.L. (2019). Multi-view Semi-supervised 3D Whole Brain Segmentation with a Self-ensemble Network. In Medical Image Computing and Computer Assisted Intervention \u2013 MICCAI 2019 (pp. 256\u2013265). Springer International Publishing. |\n| scan_2019 | McKinley, R., Rebsamen, M., Meier, R., & Wiest, R. (2020). Triplanar Ensemble of 3D-to-2D CNNs with Label-Uncertainty for Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (pp. 379\u2013387). Springer International Publishing. |\n| isen-20 | Isensee, F., J\u00e4ger, P. F., Full, P. M., Vollmuth, P., & Maier-Hein, K. H. (2021). nnU-Net for brain tumor segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, (pp. 118-132). Springer International Publishing. |\n| hnfnetv1-20 | Jia, H., Cai, W., Huang, H., & Xia, Y. (2021). H^ 2 2 NF-Net for Brain Tumor Segmentation Using Multimodal MR Imaging: 2nd Place Solution to BraTS Challenge 2020 Segmentation Task. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, (pp. 58-68). Springer International Publishing. |\n| yixinmpl-20 | Wang, Y., Zhang, Y., Hou, F., Liu, Y., Tian, J., Zhong, C., ... & He, Z. (2021). Modality-pairing learning for brain tumor segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020,(pp. 230-240). Springer International Publishing. |\n| sanet0-20 | Yuan, Y. (2021). Automatic brain tumor segmentation with scale attention network. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries BrainLes 2020, (pp. 285-294). Springer International Publishing. |\n| scan-20 & scan_lite-20 | McKinley, R., Rebsamen, M., D\u00e4twyler, K., Meier, R., Radojewski, P., & Wiest, R. (2021). Uncertainty-driven refinement of tumor-core segmentation using 3d-to-2d networks with label uncertainty. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, (pp. 401-411). Springer International Publishing. |\n\n\n## Usage\nBrainLes features [Jupyter notebook tutorials](https://github.com/BrainLesion/tutorials/tree/main/BraTS-Toolkit) for glioma segmentation with BraTS Toolkit.\nFurther, have a look at the Python examples in this repository for preprocessing (`0_preprocessing_single.py`), segmentation (`1_segmentation.py`) and fusion (`2_fusion.py`).\n\n## Brats Segmentor\nBraTS Segmentor enables orchestration of BraTS brain tumor segmentation algorithms for generation of fully-automated segmentations.\nFor segmentation, your files should be `preprocessed`, meaning they should be co-registered and skullstripped in SRI-24 space.\nYou can preprocess your files using [BrainLes preprocessing](https://github.com/BrainLesion/preprocessing) (recommended) or the deprecated preprocessor of BraTS Toolkit (see below).\n\n### Python package\nPlease have a look at `1_segmentation.py` in this repository for a demo application.\n\n### Command Line Interface (CLI)\nType `brats-segment -h` after installing the Python package to see available options.\n\n## Brats Fusionator\nBraTS Fusionator can combine the resulting candidate segmentations into consensus segmentations using fusion methods such as majority voting and iterative SIMPLE fusion.\n### Python package\nPlease have a look at `2_fusion.py` in this repository for a demo application.\n\n## Brats Preprocessor (deprecated)\nBraTS Preprocessor facilitates data standardization and preprocessing for researchers and clinicians alike. It covers the entire image analysis workflow prior to tumor segmentation, from image conversion and registration to brain extraction.\n\n> WARNING: BraTS Preprocessor is deprecated. It still works, but we recommended using [BrainLes preprocessing](https://github.com/BrainLesion/preprocessing) instead, which offers much more flexibility.\n\n### Processing Modi\nBraTS Preprocessor offers the following preprocessing modes:\n#### GPU brain extraction: \"gpu\"\nActivates HD-BET in CUDA mode. Only supported when using NVIDIA docker (Linux operating systems only for the moment).\n#### CPU brain extraction: \"cpu\"\nActivates HD-BET or ROBEX in CPU mode depending on available RAM.\n#### ROBEX brain extraction: \"robex\"\nActivates Robex brain extraction.\n#### GPU defacing: \"gpu_defacing\"\nActivates GPU defacing - not implemented yet falls back to `cpu_defacing` for the moment. \n#### CPU defacing: \"cpu_defacing\"\nActivates mri-deface to deface using CPU.\n\n### Single vs. batch processing\n BraTS preprocessor offers processing files in `batch` or `single` mode. Please have a look at the respective example scripts / CLI commands for information how to use them.\n \n For faster computation, we strictly recommend the `batch` processing mode, which avoids the additional overhead of spawning and shutting down multiple docker containers and instead does all the processing in one container. \n\n### Python package\nPlease have a look at `0_preprocessing_batch.py` and `0_preprocessing_single.py` in this repository for a demo application. You can download the example data by cloning this repository.\n\n### Command Line Interface (CLI)\n#### single file processing\nType `brats-preprocess -h` after installing the Python package to see available options.\n#### batch file processing\nType `brats-batch-preprocess -h` after installing the Python package to see available options.\n\n### Graphical User Interface (GUI)\nYou can find instructions to use and download the GUI variant here: https://neuronflow.github.io/BraTS-Preprocessor/\n> WARNING: The GUI is not nicely maintained. We encourage you to use the Python package or the CLI instead.\n\n\n### Command Line Interface (CLI)\nType `brats-fuse -h` after installing the Python package to see available options.\n\n## Contact / Feedback / Questions\nOpen an issue in this git repository or contact us via email.\n\n",
"bugtrack_url": null,
"license": "AGPL-3.0",
"summary": "BraTS Toolkit is a holistic approach to brain tumor segmentation allowing to build modular pipeliens for preprocessing, segmentation and fusion of segmentations.",
"version": "1.0.17",
"project_urls": {
"Documentation": "https://brats-toolkit.readthedocs.io/en/latest/",
"Homepage": "https://github.com/neuronflow/BraTS-Toolkit",
"Repository": "https://github.com/neuronflow/BraTS-Toolkit"
},
"split_keywords": [
"brain tumor",
" glioma",
" brats",
" segmentation",
" fusion",
" skullstripping",
" brain extraction"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "c3b2d4bf1f8e4e674f93225e4d9d9c9545bbb5ee066106dc85fea83a1975f818",
"md5": "32d10c6669a979530c8f3bcc847af988",
"sha256": "6708ddfdc23af894e261a5e17751b421519c9916fb2110924c7695b7067a19b1"
},
"downloads": -1,
"filename": "brats_toolkit-1.0.17-py3-none-any.whl",
"has_sig": false,
"md5_digest": "32d10c6669a979530c8f3bcc847af988",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": "<4.0,>=3.10",
"size": 44203,
"upload_time": "2024-09-26T12:13:18",
"upload_time_iso_8601": "2024-09-26T12:13:18.868266Z",
"url": "https://files.pythonhosted.org/packages/c3/b2/d4bf1f8e4e674f93225e4d9d9c9545bbb5ee066106dc85fea83a1975f818/brats_toolkit-1.0.17-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-09-26 12:13:18",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "neuronflow",
"github_project": "BraTS-Toolkit",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "brats-toolkit"
}
