# ProcessPLS
An Implementation of ProcessPLS in Python
[](https://doi.org/10.5281/zenodo.7074754)
## Code Writter
Implementation by Sin Yong Teng. Radboud University Nijmegen, the Netherlands.
## Implementation
In this code implementation, the sklearn syntax is used. Furthermore, the ProcessPLS algorithm has been made to be represented in directed graphs data structure. This allows for more flexibility to be used with graph theory routines.
# Functions
## Install the library
```bat
pip install processPLS
```
## Get the data
```python
from processPLS.model import *
from processPLS.datasets import *
X,Y,matrix=ValdeLoirData() #Get the data conviniently
```
## Alternatively, you can import the data yourself like this:
```python
df=pd.read_csv(r'.\ValdeLoirData.csv')
df=df.drop(columns=df.columns[0])
smell_at_rest=df.iloc[:,:5]
view=df.iloc[:,5:8]
smell_after_shaking=df.iloc[:,8:18]
tasting=df.iloc[:,18:27]
global_quality=df.iloc[:,27]
X={
'Smell at Rest':smell_at_rest,
"View":view,
"Smell after Shaking":smell_after_shaking,
"Tasting":tasting,
}
Y={"Global Quality":global_quality}
matrix = pd.DataFrame(
[
[0,0,0,0,0],
[1,0,0,0,0],
[1,1,0,0,0],
[1,1,1,0,0],
[1,1,1,1,0],
],
index=list(X.keys())+list(Y.keys()),
columns=list(X.keys())+list(Y.keys())
)
```
## Call and Fit the Process PLS model
```python
import matplotlib.pyplot as plt
model = ProcessPLS()
model.fit(X,Y,matrix)
model.plot()
plt.show()
```
## Main Function Arguments
```python
Process_PLS(cv=RepeatedKFold(n_splits=5,n_repeats=2,random_state=999),scoring='neg_mean_squared_error',max_lv=30,overwrite_lv=False,inner_forced_lv=None,outer_forced_lv=None,name=None)
'''
This function sets up the processPLS model.
cv= cross validation method (follows sklearn syntax)
scoring= loss function/ scoring function (follows sklearn syntax)
max_lv= maximum numbers of latent variable (lv) for all SIMPLS models within ProcessPLS
overwrite_LV= (True/False) A boolean to set whether inner_forced_lv and outer_forced_lv should be used instead of automatically selecting latent variables
inner_forced_lv= (dict) a specific key value combination of number of LVs to forced into the inner model. Argument overwrite_LV must be set to True for this to be used. Example input:
inner_forced_lv={
'Smell at Rest':None,
"View":3,
"Smell after Shaking":6,
"Tasting":8,
"Global Quality":13
}
inner_forced_lv= (dict) a specific key value combination of number of LVs to forced into the outer model. Argument overwrite_LV must be set to True for this to be used. Example input:
outer_forced_lv={
'Smell at Rest':3,
"View":3,
"Smell after Shaking":2,
"Tasting":5,
"Global Quality":3
}
name: (string) Optional name of model.
'''
ValdeLoirData(original=False)
'''
This function gets the data for Valde Loir Dataset
original==False: The function returns X (dataframe in dict), Y (dataframe dict), and matrix (dataframe). matrix is the adjacency matrix for the graph connections.
original==True: The function returns the raw data (dataframe) with both X and Y combined within
'''
```
## Inference/ Prediction for New Data
```python
y_pred= model.predict(Xnew)
```
# Colab Example [Here](https://colab.research.google.com/drive/1tEW7zRytpWzDoLw95N783bAAvEUhzKvX?usp=sharing)
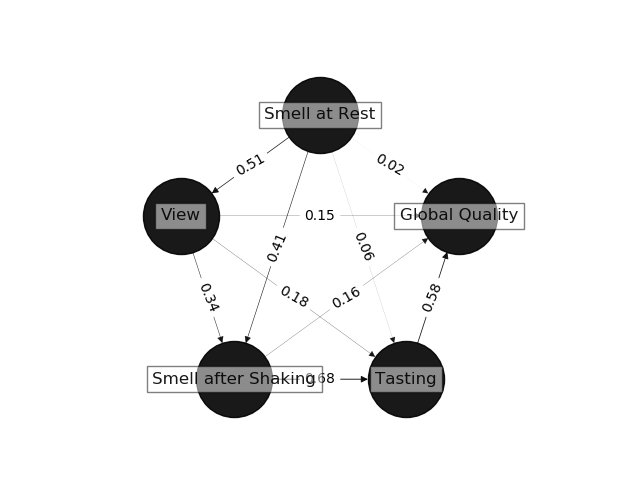
## Reproducibility
This implementation provides exactly the same output as the MATLAB version of ProcessPLS.
## Reference to Original Paper:
van Kollenburg, G., Bouman, R., Offermans, T., Gerretzen, J., Buydens, L., van Manen, H.J. and Jansen, J., 2021. Process PLS: Incorporating substantive knowledge into the predictive modelling of multiblock, multistep, multidimensional and multicollinear process data. Computers & Chemical Engineering, 154, p.107466.
For MATLAB Implementation, see this repository written by Tim Offermans.
https://gitlab.science.ru.nl/toffermans/matlab-process-pls/-/tree/main/
## How to cite this software
S.Y. Teng. (2022). tsyet12/ProcessPLS:An Implementation of ProcessPLS in Python, Zenodo Release (zenodo). Zenodo. https://doi.org/10.5281/zenodo.7074754
Raw data
{
"_id": null,
"home_page": "",
"name": "ProcessPLS",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "Path Modelling,Chemometrics,Process Analytical Technology,Machine Learning",
"author": "Sin Yong Teng",
"author_email": "tsyet12@gmail.com",
"download_url": "https://files.pythonhosted.org/packages/56/cf/74c9156b779fbe5a200fd549b5d48cd51032e7df104fe9f54287f898e104/ProcessPLS-1.9.tar.gz",
"platform": null,
"description": "# ProcessPLS\r\nAn Implementation of ProcessPLS in Python\r\n[](https://doi.org/10.5281/zenodo.7074754)\r\n\r\n\r\n## Code Writter\r\nImplementation by Sin Yong Teng. Radboud University Nijmegen, the Netherlands.\r\n\r\n## Implementation\r\nIn this code implementation, the sklearn syntax is used. Furthermore, the ProcessPLS algorithm has been made to be represented in directed graphs data structure. This allows for more flexibility to be used with graph theory routines. \r\n\r\n# Functions\r\n\r\n## Install the library\r\n```bat\r\npip install processPLS\r\n```\r\n\r\n## Get the data\r\n```python\r\nfrom processPLS.model import *\r\nfrom processPLS.datasets import *\r\nX,Y,matrix=ValdeLoirData() #Get the data conviniently\r\n```\r\n\r\n## Alternatively, you can import the data yourself like this:\r\n```python\r\ndf=pd.read_csv(r'.\\ValdeLoirData.csv')\r\ndf=df.drop(columns=df.columns[0])\r\nsmell_at_rest=df.iloc[:,:5]\r\nview=df.iloc[:,5:8]\r\nsmell_after_shaking=df.iloc[:,8:18]\r\ntasting=df.iloc[:,18:27]\r\nglobal_quality=df.iloc[:,27]\r\n\r\nX={\r\n'Smell at Rest':smell_at_rest,\r\n\"View\":view,\r\n\"Smell after Shaking\":smell_after_shaking,\r\n\"Tasting\":tasting,\r\n}\r\n\r\nY={\"Global Quality\":global_quality}\r\n\r\nmatrix = pd.DataFrame(\r\n[\r\n[0,0,0,0,0], \r\n[1,0,0,0,0],\r\n[1,1,0,0,0],\r\n[1,1,1,0,0],\r\n[1,1,1,1,0],\r\n],\r\nindex=list(X.keys())+list(Y.keys()),\r\ncolumns=list(X.keys())+list(Y.keys())\r\n)\r\n\r\n```\r\n\r\n## Call and Fit the Process PLS model\r\n```python\r\nimport matplotlib.pyplot as plt\r\nmodel = ProcessPLS()\r\nmodel.fit(X,Y,matrix)\r\nmodel.plot()\r\nplt.show()\r\n```\r\n\r\n## Main Function Arguments\r\n```python\r\nProcess_PLS(cv=RepeatedKFold(n_splits=5,n_repeats=2,random_state=999),scoring='neg_mean_squared_error',max_lv=30,overwrite_lv=False,inner_forced_lv=None,outer_forced_lv=None,name=None)\r\n\r\n'''\r\nThis function sets up the processPLS model.\r\n\r\ncv= cross validation method (follows sklearn syntax)\r\n\r\nscoring= loss function/ scoring function (follows sklearn syntax)\r\n\r\nmax_lv= maximum numbers of latent variable (lv) for all SIMPLS models within ProcessPLS\r\n\r\noverwrite_LV= (True/False) A boolean to set whether inner_forced_lv and outer_forced_lv should be used instead of automatically selecting latent variables\r\n\r\ninner_forced_lv= (dict) a specific key value combination of number of LVs to forced into the inner model. Argument overwrite_LV must be set to True for this to be used. Example input:\r\n inner_forced_lv={\r\n 'Smell at Rest':None,\r\n \"View\":3,\r\n \"Smell after Shaking\":6,\r\n \"Tasting\":8,\r\n \"Global Quality\":13\r\n }\r\n \r\n inner_forced_lv= (dict) a specific key value combination of number of LVs to forced into the outer model. Argument overwrite_LV must be set to True for this to be used. Example input:\r\n\r\n outer_forced_lv={\r\n 'Smell at Rest':3,\r\n \"View\":3,\r\n \"Smell after Shaking\":2,\r\n \"Tasting\":5,\r\n \"Global Quality\":3\r\n }\r\n \r\nname: (string) Optional name of model.\r\n\r\n'''\r\n\r\nValdeLoirData(original=False)\r\n\r\n'''\r\nThis function gets the data for Valde Loir Dataset\r\n\r\noriginal==False: The function returns X (dataframe in dict), Y (dataframe dict), and matrix (dataframe). matrix is the adjacency matrix for the graph connections.\r\n\r\noriginal==True: The function returns the raw data (dataframe) with both X and Y combined within\r\n\r\n\r\n'''\r\n\r\n```\r\n\r\n## Inference/ Prediction for New Data\r\n\r\n```python\r\ny_pred= model.predict(Xnew)\r\n\r\n```\r\n\r\n# Colab Example [Here](https://colab.research.google.com/drive/1tEW7zRytpWzDoLw95N783bAAvEUhzKvX?usp=sharing)\r\n\r\n\r\n\r\n## Reproducibility\r\nThis implementation provides exactly the same output as the MATLAB version of ProcessPLS.\r\n\r\n\r\n\r\n\r\n\r\n## Reference to Original Paper:\r\nvan Kollenburg, G., Bouman, R., Offermans, T., Gerretzen, J., Buydens, L., van Manen, H.J. and Jansen, J., 2021. Process PLS: Incorporating substantive knowledge into the predictive modelling of multiblock, multistep, multidimensional and multicollinear process data. Computers & Chemical Engineering, 154, p.107466.\r\n\r\nFor MATLAB Implementation, see this repository written by Tim Offermans.\r\nhttps://gitlab.science.ru.nl/toffermans/matlab-process-pls/-/tree/main/\r\n\r\n\r\n## How to cite this software\r\n\r\nS.Y. Teng. (2022). tsyet12/ProcessPLS:An Implementation of ProcessPLS in Python, Zenodo Release (zenodo). Zenodo. https://doi.org/10.5281/zenodo.7074754\r\n\r\n\r\n",
"bugtrack_url": null,
"license": "BSD 2-Clause",
"summary": "Implementation of ProcessPLS in Python",
"version": "1.9",
"project_urls": null,
"split_keywords": [
"path modelling",
"chemometrics",
"process analytical technology",
"machine learning"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "56cf74c9156b779fbe5a200fd549b5d48cd51032e7df104fe9f54287f898e104",
"md5": "31c1270c9843703a4a4f7bc5d94edf3c",
"sha256": "03f1f2d1b3d3a24406d06afee77abca84fd9681c4dae431a44997d5add166d19"
},
"downloads": -1,
"filename": "ProcessPLS-1.9.tar.gz",
"has_sig": false,
"md5_digest": "31c1270c9843703a4a4f7bc5d94edf3c",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 257173,
"upload_time": "2023-07-13T12:18:30",
"upload_time_iso_8601": "2023-07-13T12:18:30.323462Z",
"url": "https://files.pythonhosted.org/packages/56/cf/74c9156b779fbe5a200fd549b5d48cd51032e7df104fe9f54287f898e104/ProcessPLS-1.9.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-07-13 12:18:30",
"github": false,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"lcname": "processpls"
}