# Single Molecule Light Field Microscopy Reconstruction (PySMLFM)
[](https://github.com/Photometrics/PySMLFM/releases "GitHub stable release")
[](https://github.com/Photometrics/PySMLFM/releases "GitHub latest release")
[](https://pypi.org/project/PySMLFM/ "PyPI release")
[](https://test.pypi.org/project/PySMLFM/ "TestPyPI release")
[](https://github.com/Photometrics/PySMLFM/actions/workflows/tests.yml "Tests status")
[](https://python.org "Python versions")
[](http://isitmaintained.com/project/Photometrics/PySMLFM "Average time to resolve an issue")
[](http://isitmaintained.com/project/Photometrics/PySMLFM "Percentage of issues still open")
---
A Python solution inspired by MATLAB scripts in
[hexSMLFM](https://github.com/TheLeeLab/hexSMLFM) project.
> This 3D reconstruction code accompanies the preprint titled
"[High-density volumetric super-resolution microscopy](
https://www.biorxiv.org/content/10.1101/2023.05.02.539032v1)".
In brief, 2D localisation data (x,y) captured using a (square or hexagonal)
fourier light field microscope are turned into 3D localisations (x,y,z).
It does this by first assigning (x,y) to (x,y,u,v) space and using microscope
parameters to calculate z position via parallax. For more information, see:
[R. R. Sims, *et al.* Optica, 2020, 7, 1065](https://doi.org/10.1364/OPTICA.397172).
## Getting Started
Although this project is pure Python, one of its optional features talks to
[ImageJ/Fiji](https://imagej.net/) via [PyImageJ](https://github.com/imagej/pyimagej)
wrapper and uses PeakFit from [GDSC SMLM2](https://imagej.net/plugins/gdsc-smlm2)
plugin. That basically defines main requirements as well as restrictions for
this project.
If you don't plan to use PeakFit directly from this project, feel free to
configure your Python environment in a way you are used to. You can still
follow the instruction below, just skip installation of PyImageJ and OpenJDK.
Following instructions assume you don't have Python installed yet and focuses to
Windows platform only.
## Initial Setup
Initial setup follows PyImageJ instructions.
PyImageJ should be installed using [Conda](https://conda.io/) +
[Mamba](https://mamba.readthedocs.io/).
You can also `pip install pyimagej`, but will then need to install
[OpenJDK](https://en.wikipedia.org/wiki/OpenJDK) and
[Maven](https://maven.apache.org/) manually.
1. Install [Miniforge3](https://github.com/conda-forge/miniforge#miniforge3)
to get whole Python environment. When asked how to install, select _Just me_,
because _All users_ option would only complicate system-wide installation.
If you really need to share the installation with other users, select
location everybody can access and manually copy there also a Start menu
shortcut to Miniforge prompt created by installer for you only.
2. Run Miniforge Prompt from Start menu. It opens in your home folder by default
(e.g. `C:\Users\Me`).
3. Create new isolated virtual environment for this project (with name _MyEnv_)
to not clutter dependencies with your existing or new projects:
```
(base) C:\Users\Me> mamba create -n MyEnv -c conda-forge python=3.8 pyimagej openjdk=8
```
This command will install PyImageJ with OpenJDK 8 in new virt. env. using
Python 3.8 (the minimal Python version required).<br>
If you don't want PyImageJ, remove last two package names from the command
to get clean environment.<br>
Adjust Python version if needed.
> Note:<br>
> If you have an AMD processor and experience low performance compared to
> similar Intel CPU, installing an alternative BLAS library could help.
> For example, to initialize new environment with BLIS library run:
> ```
> (base) C:\Users\Me> mamba create -n MyEnv -c conda-forge python=3.8 pyimagej openjdk=8 blas=*=blis
> ```
4. **Whenever you want to use _MyEnv_ environment, activate it first**:
```
(base) C:\Users\Me> mamba activate MyEnv
```
5. Ensure conda-forge is the first update channel:
```
(MyEnv) C:\Users\Me> conda config --add channels conda-forge
(MyEnv) C:\Users\Me> conda config --set channel_priority strict
```
6. Download [Fiji](https://imagej.net/software/fiji/) from official source,
unpack it, run `ImageJ-win64.exe` and install '_GDSC SMLM2_' plugin via
_Help_‑>_Update..._ menu.
## Install PySMLFM
Install either stable release or development version as shown below.
### Latest Stable Release
- Install the package from Python Package Index (PyPI):
```
(MyEnv) C:\Users\Me> pip install PySMLFM
```
### Latest Development Version
- Get the ZIP archive with sources from GitHub (`PySMLFM-main.zip`).
- Install the package from source archive extracted e.g. in your home folder
(`C:\Users\Me\PySMLFM-main`):
```
(MyEnv) C:\Users\Me> cd PySMLFM-main
(MyEnv) C:\Users\Me\PySMLFM-main> pip install .
```
## Execution
There are two ways how to run an application installed by this project -
by running a Python command from Miniforge Prompt or via installed `.exe`
helper script.
### Via Python Command
Run the command line application:
```
(MyEnv) C:\Users\Me> python -m smlfm_cli
```
or the GUI application:
```
(MyEnv) C:\Users\Me> python -m smlfm_gui
```
### Via Helper Script
During the installation is created a helper script that simplifies the execution
of the application. It is a self-extracting ZIP file that executes similar
command as above but uses for it the `python.exe` from the virt. env. where it
was created from. Because of that, the EXE helper works only on the that
computer and cannot be shared with anyone else, but it can be copied anywhere on
your computer.
Simply run it with:
```
(MyEnv) C:\Users\Me> smlfm-cli
```
or:
```
(MyEnv) C:\Users\Me> smlfm-gui
```
Notice the dash in the command name, it is not an underscore.
## Usage
If you already tried running the CLI application, you saw an error message.
It is because the CLI application requires one or two options as input.
### Configuration
The type of the input option is detected automatically from given file extension:
- A configuration with `.json` file extension.<br>
It contains all possible parameters for the localisation process, output
location, graphs to show, etc. If not specified, a default configuration file
is used. As a starting point, make your own copy and modify the parameters
accordingly. See
`C:\Users\Me\miniforge\envs\MyEnv\Lib\site-packages\smlfm\data\default-config.json`.
- A 2D localisations with `.csv` or `.xls` file extension.<br>
If not specified on command line, the path to the file must be set in the
configuration file under `csv_file` key.<br>
By default the CSV is expected in the format generated by PeakFit plugin
in ImageJ/Fiji. A few other formats are supported, namely Thunderstorm and
Picasso. Adjust the configuration file key `csv_format` where the value is
all in upper-case.
For example, run the application with customized configuration and data, all
stored `C:\MyExperiment` folder:
```
(MyEnv) C:\MyExperiment> python -m smlfm_cli my-config.json my-localisations.csv
```
For quick access to the EXE helper script without opening command prompt you can
create a shortcut on the desktop or in your experiment folder.
Open the path with `smlfm-cli.exe` in File Explorer (by default it is in
`C:\Users\Me\miniforge\envs\MyEnv\Scripts\ `), drag the EXE file with
mouse and drop it on the desktop or target folder while holding the `Alt` key.<br>
The shortcut has working directory set by default to the folder with the EXE file.
It is recommended changing it to a location where you want the results to be
stored. Right-click on the shortcut, select _Properties_ and modify '_Start in:_'
box accordingly. Also don't forget to add the input options at the end of the
command in the '_Target:_' box. Now you can double-click the shortcut anytime
you want to re-run the 3D localisation.
### Processing
With valid configuration the 2D localisation data is loaded from CSV file and
every localisation point is assigned to the nearest lens from micro-lens array.
With default configuration the localisations and lens centres are shown in form
of 2D graph. The user can visually check the alignment. To continue, the graph
window must be closed and then the user must either confirm the alignment or
abort the process to adjust the configuration to fix the alignment. Simply
follow the instructions in the command prompt window.
Based on the amount of input localisations and computer performance, the
processing can take from a few seconds to minutes or even hours. The command
prompt window prints a message about the progress every 1000 frames processed.
### Results
If everything completed successfully, three graphs with results are shown and
the results are stored in the disk. In the _current working directory_
(e.g. `C:\MyExperiment`) is created a subdirectory for every execution.
The subdirectory name starts with `3D Fitting ` prefix followed by a timestamp
and a CSV filename used.<br>
Each folder contains JSON configuration file used (`config.json`), calculated
3D localisations (`locs3D.csv`), an output for
[ViSP viewer](https://www.nature.com/articles/nmeth.2566) (`ViSP.3d`) and
a Python script (`figures.pyw`) that reconstructs exactly the results graphs at
any time later without a need to start the processing from beginning.
To open the saved figures run a command that takes relative path to it like this
(quote the path with spaces):
```
(MyEnv) C:\MyExperiment> python "3D Fitting - 20240213-203318 - my-localisations.csv/figures.pyw"
```
When you use `python` command like above, the prompt will be blocked until all
graphs are closed. To open the figures without blocking the prompt use `pythonw`
instead.
## Screenshots
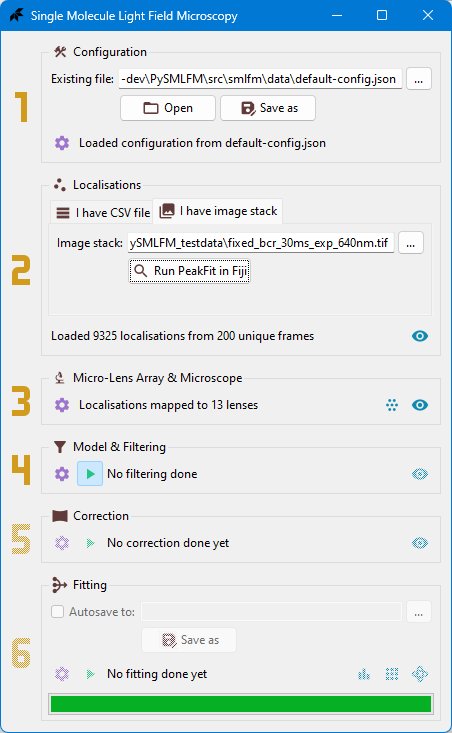
### Main Window
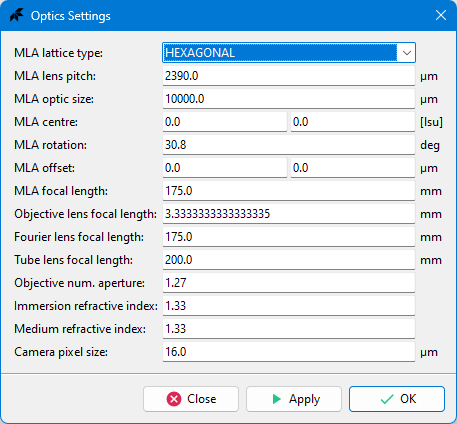
### Optics Settings Dialog
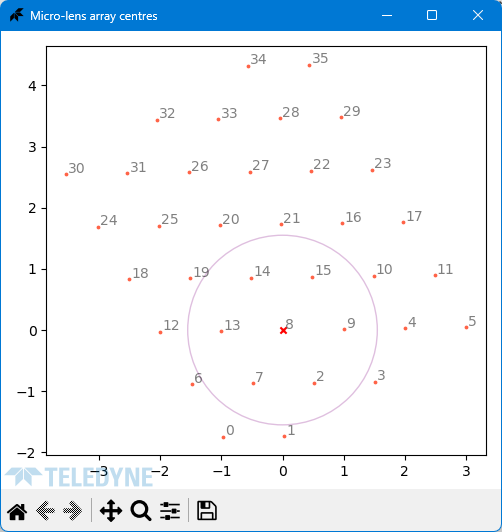
### Micro Lens Array Layout
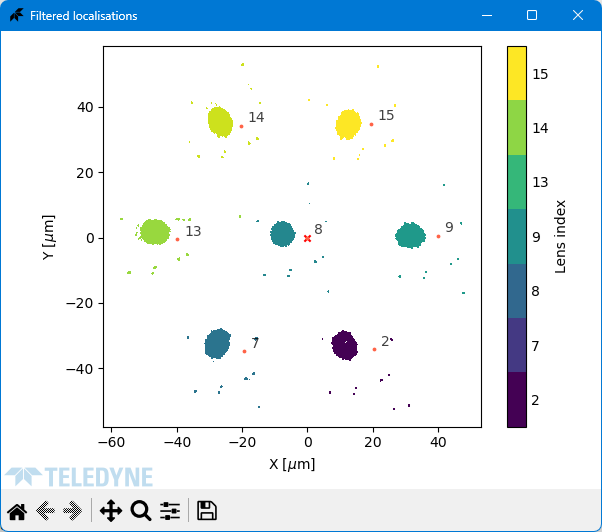
### Filtered Localisations
### 3D Reconstruction
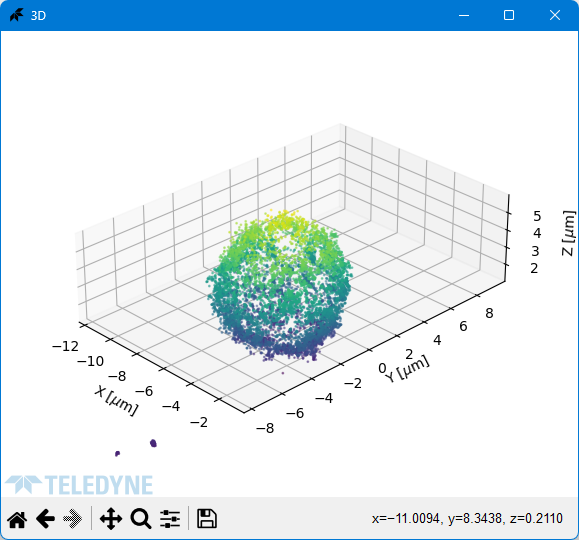
Raw data
{
"_id": null,
"home_page": "https://github.com/Photometrics/PySMLFM",
"name": "PySMLFM",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": null,
"keywords": "microscopy, lightfield",
"author": "Teledyne Photometrics",
"author_email": "Tom\u00e1\u0161 Han\u00e1k <tomas.hanak@teledyne.com>",
"download_url": "https://files.pythonhosted.org/packages/93/ea/29cd65984e359cf141464eeb6489d418c18e01322133c5eb49eb9d299348/pysmlfm-1.0.0.tar.gz",
"platform": null,
"description": "# Single Molecule Light Field Microscopy Reconstruction (PySMLFM)\n\n[](https://github.com/Photometrics/PySMLFM/releases \"GitHub stable release\")\n[](https://github.com/Photometrics/PySMLFM/releases \"GitHub latest release\") \n[](https://pypi.org/project/PySMLFM/ \"PyPI release\")\n[](https://test.pypi.org/project/PySMLFM/ \"TestPyPI release\") \n[](https://github.com/Photometrics/PySMLFM/actions/workflows/tests.yml \"Tests status\")\n[](https://python.org \"Python versions\") \n[](http://isitmaintained.com/project/Photometrics/PySMLFM \"Average time to resolve an issue\")\n[](http://isitmaintained.com/project/Photometrics/PySMLFM \"Percentage of issues still open\")\n---\n\nA Python solution inspired by MATLAB scripts in\n[hexSMLFM](https://github.com/TheLeeLab/hexSMLFM) project.\n\n> This 3D reconstruction code accompanies the preprint titled\n\"[High-density volumetric super-resolution microscopy](\nhttps://www.biorxiv.org/content/10.1101/2023.05.02.539032v1)\".\nIn brief, 2D localisation data (x,y) captured using a (square or hexagonal)\nfourier light field microscope are turned into 3D localisations (x,y,z).\nIt does this by first assigning (x,y) to (x,y,u,v) space and using microscope\nparameters to calculate z position via parallax. For more information, see:\n[R. R. Sims, *et al.* Optica, 2020, 7, 1065](https://doi.org/10.1364/OPTICA.397172).\n\n## Getting Started\n\nAlthough this project is pure Python, one of its optional features talks to\n[ImageJ/Fiji](https://imagej.net/) via [PyImageJ](https://github.com/imagej/pyimagej)\nwrapper and uses PeakFit from [GDSC SMLM2](https://imagej.net/plugins/gdsc-smlm2)\nplugin. That basically defines main requirements as well as restrictions for\nthis project.\n\nIf you don't plan to use PeakFit directly from this project, feel free to\nconfigure your Python environment in a way you are used to. You can still\nfollow the instruction below, just skip installation of PyImageJ and OpenJDK.\n\nFollowing instructions assume you don't have Python installed yet and focuses to\nWindows platform only.\n\n## Initial Setup\n\nInitial setup follows PyImageJ instructions.\n\nPyImageJ should be installed using [Conda](https://conda.io/) +\n[Mamba](https://mamba.readthedocs.io/).\nYou can also `pip install pyimagej`, but will then need to install\n[OpenJDK](https://en.wikipedia.org/wiki/OpenJDK) and\n[Maven](https://maven.apache.org/) manually.\n\n1. Install [Miniforge3](https://github.com/conda-forge/miniforge#miniforge3)\n to get whole Python environment. When asked how to install, select _Just me_,\n because _All users_ option would only complicate system-wide installation.\n If you really need to share the installation with other users, select\n location everybody can access and manually copy there also a Start menu\n shortcut to Miniforge prompt created by installer for you only.\n2. Run Miniforge Prompt from Start menu. It opens in your home folder by default\n (e.g. `C:\\Users\\Me`).\n3. Create new isolated virtual environment for this project (with name _MyEnv_)\n to not clutter dependencies with your existing or new projects:\n ```\n (base) C:\\Users\\Me> mamba create -n MyEnv -c conda-forge python=3.8 pyimagej openjdk=8\n ```\n This command will install PyImageJ with OpenJDK 8 in new virt. env. using\n Python 3.8 (the minimal Python version required).<br>\n If you don't want PyImageJ, remove last two package names from the command\n to get clean environment.<br>\n Adjust Python version if needed.\n\n > Note:<br>\n > If you have an AMD processor and experience low performance compared to\n > similar Intel CPU, installing an alternative BLAS library could help.\n > For example, to initialize new environment with BLIS library run:\n > ```\n > (base) C:\\Users\\Me> mamba create -n MyEnv -c conda-forge python=3.8 pyimagej openjdk=8 blas=*=blis\n > ```\n4. **Whenever you want to use _MyEnv_ environment, activate it first**:\n ```\n (base) C:\\Users\\Me> mamba activate MyEnv\n ```\n5. Ensure conda-forge is the first update channel:\n ```\n (MyEnv) C:\\Users\\Me> conda config --add channels conda-forge\n (MyEnv) C:\\Users\\Me> conda config --set channel_priority strict\n ```\n6. Download [Fiji](https://imagej.net/software/fiji/) from official source,\n unpack it, run `ImageJ-win64.exe` and install '_GDSC SMLM2_' plugin via\n _Help_‑>_Update..._ menu.\n\n## Install PySMLFM\n\nInstall either stable release or development version as shown below.\n\n### Latest Stable Release\n\n- Install the package from Python Package Index (PyPI):\n ```\n (MyEnv) C:\\Users\\Me> pip install PySMLFM\n ```\n\n### Latest Development Version\n\n- Get the ZIP archive with sources from GitHub (`PySMLFM-main.zip`).\n- Install the package from source archive extracted e.g. in your home folder\n (`C:\\Users\\Me\\PySMLFM-main`):\n ```\n (MyEnv) C:\\Users\\Me> cd PySMLFM-main\n (MyEnv) C:\\Users\\Me\\PySMLFM-main> pip install .\n ```\n\n## Execution\n\nThere are two ways how to run an application installed by this project -\nby running a Python command from Miniforge Prompt or via installed `.exe`\nhelper script.\n\n### Via Python Command\n\nRun the command line application:\n```\n(MyEnv) C:\\Users\\Me> python -m smlfm_cli\n```\nor the GUI application:\n```\n(MyEnv) C:\\Users\\Me> python -m smlfm_gui\n```\n\n### Via Helper Script\n\nDuring the installation is created a helper script that simplifies the execution\nof the application. It is a self-extracting ZIP file that executes similar\ncommand as above but uses for it the `python.exe` from the virt. env. where it\nwas created from. Because of that, the EXE helper works only on the that\ncomputer and cannot be shared with anyone else, but it can be copied anywhere on\nyour computer.\n\nSimply run it with:\n```\n(MyEnv) C:\\Users\\Me> smlfm-cli\n```\nor:\n```\n(MyEnv) C:\\Users\\Me> smlfm-gui\n```\nNotice the dash in the command name, it is not an underscore.\n\n## Usage\n\nIf you already tried running the CLI application, you saw an error message.\nIt is because the CLI application requires one or two options as input.\n\n### Configuration\n\nThe type of the input option is detected automatically from given file extension:\n- A configuration with `.json` file extension.<br>\n It contains all possible parameters for the localisation process, output\n location, graphs to show, etc. If not specified, a default configuration file\n is used. As a starting point, make your own copy and modify the parameters\n accordingly. See\n `C:\\Users\\Me\\miniforge\\envs\\MyEnv\\Lib\\site-packages\\smlfm\\data\\default-config.json`.\n- A 2D localisations with `.csv` or `.xls` file extension.<br>\n If not specified on command line, the path to the file must be set in the\n configuration file under `csv_file` key.<br>\n By default the CSV is expected in the format generated by PeakFit plugin\n in ImageJ/Fiji. A few other formats are supported, namely Thunderstorm and\n Picasso. Adjust the configuration file key `csv_format` where the value is\n all in upper-case.\n\nFor example, run the application with customized configuration and data, all\nstored `C:\\MyExperiment` folder:\n ```\n (MyEnv) C:\\MyExperiment> python -m smlfm_cli my-config.json my-localisations.csv\n ```\n\nFor quick access to the EXE helper script without opening command prompt you can\ncreate a shortcut on the desktop or in your experiment folder.\nOpen the path with `smlfm-cli.exe` in File Explorer (by default it is in\n`C:\\Users\\Me\\miniforge\\envs\\MyEnv\\Scripts\\ `), drag the EXE file with\nmouse and drop it on the desktop or target folder while holding the `Alt` key.<br>\nThe shortcut has working directory set by default to the folder with the EXE file.\nIt is recommended changing it to a location where you want the results to be\nstored. Right-click on the shortcut, select _Properties_ and modify '_Start in:_'\nbox accordingly. Also don't forget to add the input options at the end of the\ncommand in the '_Target:_' box. Now you can double-click the shortcut anytime\nyou want to re-run the 3D localisation.\n\n### Processing\n\nWith valid configuration the 2D localisation data is loaded from CSV file and\nevery localisation point is assigned to the nearest lens from micro-lens array.\nWith default configuration the localisations and lens centres are shown in form\nof 2D graph. The user can visually check the alignment. To continue, the graph\nwindow must be closed and then the user must either confirm the alignment or\nabort the process to adjust the configuration to fix the alignment. Simply\nfollow the instructions in the command prompt window.\n\nBased on the amount of input localisations and computer performance, the\nprocessing can take from a few seconds to minutes or even hours. The command\nprompt window prints a message about the progress every 1000 frames processed.\n\n### Results\n\nIf everything completed successfully, three graphs with results are shown and\nthe results are stored in the disk. In the _current working directory_\n(e.g. `C:\\MyExperiment`) is created a subdirectory for every execution.\nThe subdirectory name starts with `3D Fitting ` prefix followed by a timestamp\nand a CSV filename used.<br>\nEach folder contains JSON configuration file used (`config.json`), calculated\n3D localisations (`locs3D.csv`), an output for\n[ViSP viewer](https://www.nature.com/articles/nmeth.2566) (`ViSP.3d`) and\na Python script (`figures.pyw`) that reconstructs exactly the results graphs at\nany time later without a need to start the processing from beginning.\n\nTo open the saved figures run a command that takes relative path to it like this\n(quote the path with spaces):\n```\n (MyEnv) C:\\MyExperiment> python \"3D Fitting - 20240213-203318 - my-localisations.csv/figures.pyw\"\n```\nWhen you use `python` command like above, the prompt will be blocked until all\ngraphs are closed. To open the figures without blocking the prompt use `pythonw`\ninstead.\n\n## Screenshots\n\n### Main Window\n\n\n\n### Optics Settings Dialog\n\n\n\n### Micro Lens Array Layout\n\n\n\n### Filtered Localisations\n\n\n\n### 3D Reconstruction\n\n\n",
"bugtrack_url": null,
"license": "GPL-3.0-or-later",
"summary": "Single Molecule Light Field Microscopy Reconstruction",
"version": "1.0.0",
"project_urls": {
"Homepage": "https://github.com/Photometrics/PySMLFM",
"download": "https://pypi.org/project/PySMLFM/",
"source": "https://github.com/Photometrics/PySMLFM",
"tracker": "https://github.com/Photometrics/PySMLFM/issues"
},
"split_keywords": [
"microscopy",
" lightfield"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "84c0cf2b787439e82e0eb0a99398f7bbcb18c0668271e56b17754fb96fb16417",
"md5": "a4b56207247b1ebc3b7087f53bc6cace",
"sha256": "6044c1f1a5fc943f0f98c2f7e7f51590772b40ece9ac61dc5139033ba98c48c5"
},
"downloads": -1,
"filename": "PySMLFM-1.0.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "a4b56207247b1ebc3b7087f53bc6cace",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 117915,
"upload_time": "2024-06-18T17:37:01",
"upload_time_iso_8601": "2024-06-18T17:37:01.669690Z",
"url": "https://files.pythonhosted.org/packages/84/c0/cf2b787439e82e0eb0a99398f7bbcb18c0668271e56b17754fb96fb16417/PySMLFM-1.0.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "93ea29cd65984e359cf141464eeb6489d418c18e01322133c5eb49eb9d299348",
"md5": "4aed261eeca9eb298940dce67468a309",
"sha256": "1581daec37f28ce0df3a6338a6415234362cd2ef09da404e251d5f194e0c4c39"
},
"downloads": -1,
"filename": "pysmlfm-1.0.0.tar.gz",
"has_sig": false,
"md5_digest": "4aed261eeca9eb298940dce67468a309",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 93627,
"upload_time": "2024-06-18T17:37:22",
"upload_time_iso_8601": "2024-06-18T17:37:22.677372Z",
"url": "https://files.pythonhosted.org/packages/93/ea/29cd65984e359cf141464eeb6489d418c18e01322133c5eb49eb9d299348/pysmlfm-1.0.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-06-18 17:37:22",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "Photometrics",
"github_project": "PySMLFM",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "pysmlfm"
}
