| Name | aignostics JSON |
| Version |
0.2.196
 JSON
JSON |
| download |
| home_page | None |
| Summary | 🔬 Python SDK providing access to the Aignostics Platform. Includes Aignostics Launchpad (Desktop Application), Aignostics CLI (Command-Line Interface), example notebooks, and Aignostics Client Library. |
| upload_time | 2025-10-26 23:41:37 |
| maintainer | None |
| docs_url | None |
| author | None |
| requires_python | <3.14,>=3.11 |
| license | MIT License Copyright (c) [2025] [Aignostics GmbH (support@aignostics.com)] Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software. THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE. |
| keywords |
act
aignostics
atlas
codecov
copier
cyclonedx
detect-secrets
devcontainer
dicom
digital-pathology
docker
git-cliff
image-data-commons
jupyter
machine-learning
marimo
medical-imaging
mypy
nicegui
nox
oe-python-template
openslide
pip-audit
pip-licenses
pre-commit
pydantic
pydicom
pypi
pytest
python
qupath
readthedocs
ruff
sonarcloud
sonarqube
sphinx
typer
uv
whole-slide-imaging
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
[//]: # (README.md generated from docs/partials/README_*.md)
# 🔬Aignostics Python SDK
[](https://github.com/aignostics/python-sdk/blob/main/LICENSE)
[](https://pypi.org/project/aignostics/)
[](https://github.com/aignostics/python-sdk/actions/workflows/ci-cd.yml)
[](https://aignostics.readthedocs.io/en/latest/)
[](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)
[](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)
[](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)
[](https://codecov.io/gh/aignostics/python-sdk)
[](https://aignostics.betteruptime.com)
> [!NOTE]
> The Aignostics Python SDK is in alpha, with [Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) and the [Aignostics Platform](https://platform.aignostics.com) in [early access](https://www.linkedin.com/posts/aignostics_introducing-atlas-he-tme-aignostics-is-activity-7325865745827979265-Sya9?utm_source=share&utm_medium=member_desktop&rcm=ACoAABRmV7cBCGv8eM_ot_kRTrBsb12olQvoLS4).
> Watch or star this repository to receive updates on new features and improvements of the SDK.
---
## Introduction
The **Aignostics Python SDK** includes multiple pathways to interact with the
**Aignostics Platform**:
1. Use the **Aignostics Launchpad** to analyze whole slide images with advanced computational pathology applications like
[Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) directly from your desktop.
View your results by launching popular tools such as [QuPath](https://qupath.github.io/) and Python Notebooks with one click.
The app runs on Mac OS X, Windows, and Linux.
2. Use the **Aignostics Command-line interface (CLI)** to run applications directly from your terminal or shell scripts.
THe CLI lets you query public datasets from the [NCI Image Data Commons (IDC)](https://portal.imaging.datacommons.cancer.gov/),
process both public and private whole slide images, and easily download results. The CLI is available for Mac OS X, Windows, and Linux.
3. Use the included **example notebooks** as starting points to run applications
directly from your preferred notebook environment. We support Marimo and Jupyter based notebooks environments including Google Collab.
4. Use the **Aignostics Client Library** to seamlessly integrate the Aignostics Platform with your enterprise image management systems and scientific workflows.
The client provides a simple way to access the Aignostics Platform API from your Python codebase. We support Python 3.11 and above.
### We take quality and security seriously
We know you take **quality** and **security** as seriously as we do. That's why
the Aignostics Python SDK is built following best practices and with full
transparency. This includes (1) making the complete
[source code of the SDK
available on GitHub](https://github.com/aignostics/python-sdk/), maintaining a
(2)
[A-grade code quality](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)
with [high test coverage](https://app.codecov.io/gh/aignostics/python-sdk) in
all releases, (3) achieving
[A-grade security](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)
with
[active scanning of dependencies](https://github.com/aignostics/python-sdk/issues/4),
and (4) providing
[extensive documentation](hhttps://aignostics.readthedocs.io/en/latest/). Read
more about how we achieve
[operational excellence](https://aignostics.readthedocs.io/en/latest/operational_excellence.html) and
[security](https://aignostics.readthedocs.io/en/latest/security.html).
## Quick Start
> [!Note]
> See as follows for a quick start guide to get you up and running with the Aignostics Python SDK as quickly as possible.
> If you first want to learn bout the basic concepts and components of the Aignostics Platform skip to that section below.
> The further reading section points you to reference documentation listing all available CLI commands, methods and classes provided by the client library, operations of the API, how we achieve operational excellence, security, and more.
> If you are not familiar with terminology please check the glossary at the end of this document.
### Launchpad: Run your first computational pathology analysis in 10 minutes from your desktop
The **Aignostics Launchpad** is a graphical desktop application that allows you to run
applications on whole slide images (WSIs) from your computer, and inspect results with QuPath and Python Notebooks with one click. It is designed to be user-friendly and intuitive, for use by Research Pathologists and Data Scientists.
The Launchpad is available for Mac OS X, Windows, and Linux, and can be installed easily:
1. Visit the [Quick Start](https://platform.aignostics.com/getting-started/quick-start)
page in the Aignostics Console.
2. Copy the installation script and paste it into your terminal - compatible with MacOS, Windows, and Linux.
3. Launch the application by running `uvx aignostics launchpad`.
4. Follow the intuitive graphical interface to analyze public datasets or your own whole slide images
with [Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) and other computational pathology applications.
> [!Note]
> The Launchpad features a growing ecosystem of extensions that seamlessly integrate with standard digital pathology tools. To use the Launchpad with all available extensions, run `uvx --from "aignostics[qupath,marimo]" aignostics launchpad`. Currently available extensions are:
>
> 1. **QuPath extension**: View your application results in [QuPath](https://qupath.github.io/) with a single click. The Launchpad creates QuPath projects on-the-fly.
> 2. **Marimo extension**: Analyze your application results using [Marimo](https://marimo.io/) notebooks embedded in the Launchpad. You don't have to leave the Launchpad to do real data science.
### CLI: Manage datasets and application runs from your terminal
The Python SDK includes the **Aignostics CLI**, a Command-Line Interface (CLI) that allows you to
interact with the Aignostics Platform directly from your terminal or shell script.
See as follows for a simple example where we download a sample dataset for the [Atlas
H&E-TME application](https://www.aignostics.com/products/he-tme-profiling-product), submit an application run, and download the results.
```shell
# Download a sample dataset from the NCI Image Data Commons (IDC) portal to your current working directory
# As the dataset id refers to the TCGA LUAD collection, this creates a directory tcga_luad with the DICOM files
uvx aignostics dataset idc download 1.3.6.1.4.1.5962.99.1.1069745200.1645485340.1637452317744.2.0 data/
# Prepare the metadata for the application run by creating a metadata.csv, extracting
# the required metadata from the DICOM files. We furthermore add the required
# information about the tissue type and disease. TODO (Helmut): Update
uvx aignostics application run prepare he-tme data/tcga_luad/run.csv data/
# Edit the metadata.csv to insert the required information about the staining method, tissue type and disease
# Adapt to your favourite editor
nano tcga_luad/metadata.csv
# Upload the metadata.csv and referenced whole slide images to the Aignostics Platform
uvx aignostics application run upload he-tme data/tcga_luad/run.csv
# Submit the application run and print tha run id
uvx aignostics application run submit he-tme data/tcga_luad/run.csv
# Check the status of the application run you submitted
uvx aignostics application run list
# Incrementally download results when they become available
# Fill in the id from the output in the previous step
uvx aignostics application run result download APPLICATION_RUN_ID
```
For convenience the the `application run execute` command combines preparation, upload, submission and download.
The below is equivalent to the above, while adding additionally required metadata using a mapping:
```shell
uvx aignostics dataset idc download 1.3.6.1.4.1.5962.99.1.1069745200.1645485340.1637452317744.2.0 data/
uvx aignostics application run execute he-tme data/tcga_luad/run.csv data/ ".*\.dcm:staining_method=H&E,tissue=LUNG,disease=LUNG_CANCER"
```
The CLI provides extensive help:
```shell
uvx aignostics --help # list all spaces such as application, dataset, bucket and system,
uvx aignostics application --help # list subcommands in the application space
uvx aignostics application run --help # list subcommands in the application run sub-space
uvx aignostics application run list --help # show help for specific command
uvx aignostics application run execute --help # show help for another command
```
Check out our
[CLI reference documentation](https://aignostics.readthedocs.io/en/latest/cli_reference.html)
to learn about all commands and options available.
### Example Notebooks: Interact with the Aignostics Platform from your Python Notebook environment
> [!IMPORTANT]
> Before you get started, you need to set up your authentication credentials if
> you did not yet do so! Please visit
> [your personal dashboard on the Aignostics Platform website](https://platform.aignostics.com/getting-started/quick-start)
> and follow the steps outlined in the `Use in Python Notebooks` section.
The Python SDK includes Jupyter and Marimo notebooks to help you get started interacting
with the Aignostics Platform in your notebook environment.
The notebooks showcase the interaction with the Aignostics Platform using our "Test Application". To run one them,
please follow the steps outlined in the snippet below to clone this repository and start either the
[Jupyter](https://docs.jupyter.org/en/latest/index.html)
([examples/notebook.ipynb](https://github.com/aignostics/python-sdk/blob/main/examples/notebook.ipynb))
or [Marimo](https://marimo.io/)
([examples/notebook.py](https://github.com/aignostics/python-sdk/blob/main/examples/notebook.py))
notebook:
```shell
# clone the `python-sdk` repository
git clone https://github.com/aignostics/python-sdk.git
# within the cloned repository, install the SDK and all dependencies
uv sync --all-extras
# show jupyter example notebook in the browser
uv run jupyter notebook examples/notebook.ipynb
# show marimo example notebook in the browser
uv run marimo edit examples/notebook.py
```
> [!Note]
> You can as well run a notebook within the Aignostics Launchpad. To do so, select the
> Run you want to inspect in the left sidebar, and click the button "Open in Python Notebook".
### Client Library: Call the Aignostics Platform API from your Python scripts
> [!IMPORTANT]\
> Before you get started, you need to set up your authentication credentials if
> you did not yet do so! Please visit
> [your personal dashboard on the Aignostics Platform website](https://platform.aignostics.com/getting-started/quick-start)
> and follow the steps outlined in the `Enterprise Integration` section.
Next to using the Launchpad, CLI and example notebooks, the Python SDK includes the
*Aignostics Client Library* for integration with your Python Codebase.
The following sections outline how to install the Python SDK for this purpose and
interact with the Client.
### Installation
The Aignostics Python SDK is published on the [Python Package Index (PyPI)](https://pypi.org/project/aignostics/),
is compatible with Python 3.11 and above, and can be installed via via `uv` or `pip`:
**Install with [uv](https://docs.astral.sh/uv/):** If you don't have uv
installed follow [these instructions](https://docs.astral.sh/uv/getting-started/installation/).
```shell
# Add Aignostics Python SDK as dependency to your project
uv add aignostics
```
**Install with [pip](https://pip.pypa.io/en/stable/)**
```shell
# Add Python SDK as dependency to your project
pip install aignostics
```
#### Usage
The following snippet shows how to use the Client to submit an application
run:
```python
from aignostics import platform
# initialize the client
client = platform.Client()
# submit an application run
application_run = client.runs.submit(
application_id="test-app",
items=[
platform.InputItem(
external_id="slide-1",
input_artifacts=[
platform.InputArtifact(
name="whole_slide_image",
download_url="<a signed url to download the data>",
metadata={
"checksum_base64_crc32c": "AAAAAA==",
"resolution_mpp": 0.25,
"width_px": 1000,
"height_px": 1000,
},
)
],
),
],
)
# wait for the results and download incrementally as they become available
application_run.download_to_folder("path/to/download/folder")
```
Please look at the notebooks in the `example` folder for a more detailed example
and read the
[client reference documentation](https://aignostics.readthedocs.io/en/latest/lib_reference.html)
to learn about all classes and methods.
##### Defining the input for an application run
When creating an application run, you need to specify the `application_id` and optionally the
`application_version` (version number) of the application you want to run. If you omit the version,
the latest version will be used automatically. Additionally, you need to define the input items you
want to process in the run. The input items are defined as follows:
```python
platform.InputItem(
external_id="1",
input_artifacts=[
platform.InputArtifact(
name="whole_slide_image", # defined by the application version's input artifact schema
download_url="<a signed url to download the data>",
metadata={ # defined by the application version's input artifact schema
"checksum_base64_crc32c": "N+LWCg==",
"resolution_mpp": 0.46499982,
"width_px": 3728,
"height_px": 3640,
},
)
],
),
```
For each item you want to process, you need to provide a unique `reference`
string. This is used to identify the item in the results later on. The
`input_artifacts` field is a list of `InputArtifact` objects, which defines what
data & metadata you need to provide for each item. The required artifacts depend
on the application version you want to run - in the case of test application,
there is only one artifact required, which is the image to process on. The
artifact name is defined as `whole_slide_image` for this application.
The `download_url` is a signed URL that allows the Aignostics Platform to
download the image data later during processing.
#### Self-signed URLs for large files
To make the whole slide images you want to process available to the Aignostics Platform, you
need to provide a signed URL that allows the platform to download the data.
Self-signed URLs for files in google storage buckets can be generated using the
`generate_signed_url`
([code](https://github.com/aignostics/python-sdk/blob/407e74f7ae89289b70efd86cbda59ec7414050d5/src/aignostics/client/utils.py#L85)).
**We expect that you provide the
[required credentials](https://cloud.google.com/docs/authentication/application-default-credentials)
for the Google Storage Bucket**
## Platform
### Overview
The **Aignostics Platform** is a comprehensive cloud-based service that allows organizations to leverage advanced computational pathology applications without the need for specialized expertise or complex infrastructure. Via its API it provides a standardized, secure interface for accessing Aignostics' portfolio of advanced computational pathology applications. These applications perform machine learning based tissue and cell analysis on histopathology slides, delivering quantitative measurements, visual representations, and detailed statistical data.
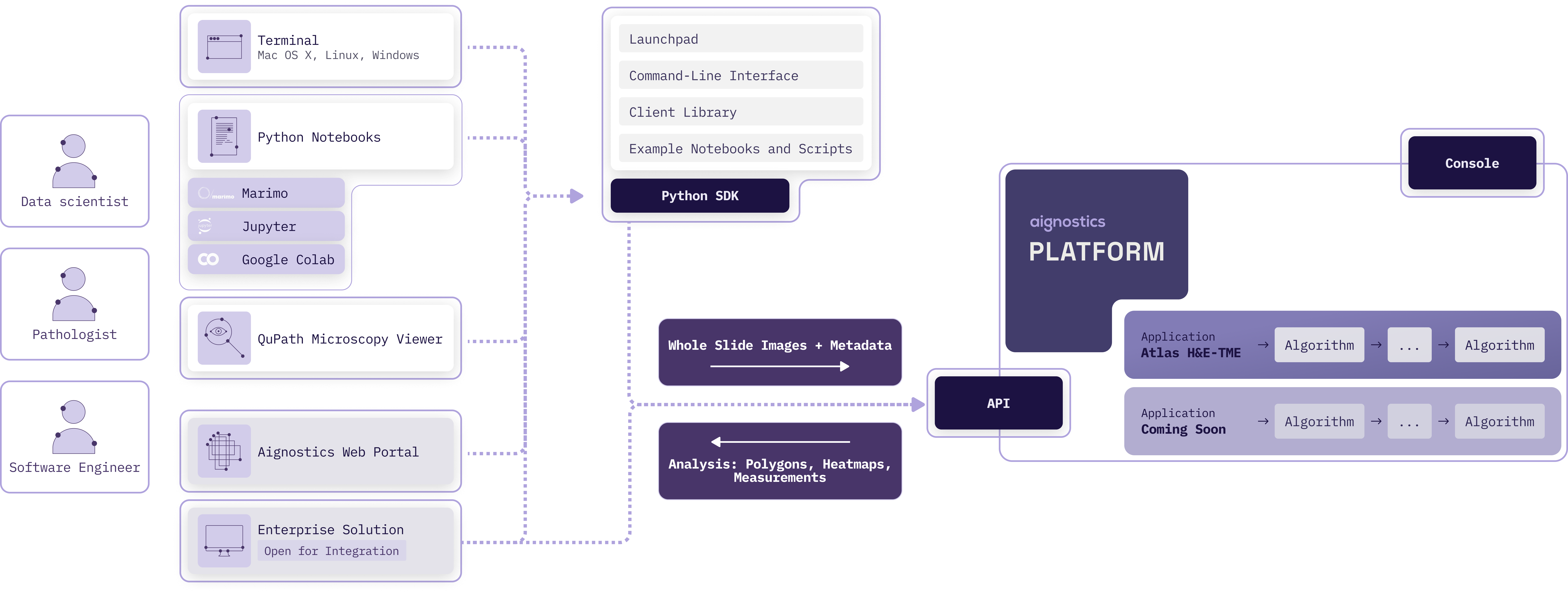
### Key Features
Aignostics Platform offers key features designed to maximize value for its users:
1. **Run Aignostics applications:** Run Aignostics advanced computational pathology applications like [Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) on your whole slide images (WSIs) and receive results in a easy to inspect formats.
2. **Multiple Access Points:** Interact with the platform via various pathways, from **Aignostics Launchpad** (desktop application for MacOS, Windows and Linux), **Aignostics CLI** (command-line interface for your terminal or shell scripts), **Example Notebooks** (we support Jupyter and Marimo), **Aignostics Client Library** (for integration with your Python codebase), or directly through the **API of the Aignostics Platform** (for integration with any programming language). Contact your business partner at Aignostics if you are interested to discuss a direct integration with your Imaging Management Systems (IMS) and Laboratory Information Management Systems (LIMS).
3. **Secure Data Handling:** Maintain control of your slide data through secure self-signed URLs. Results are automatically deleted after 30 days, and can be deleted earlier by the user.
4. **High-throughput processing with incremental results delivery:** Submit up to 500 whole slide images (WSI) in one batch request. Access results for individual slides as they completed processing, without having to wait for the entire batch to finish.
5. **Standard formats:** Support for commonly used image formats in digital pathology such as pyramidal DICOM, TIFF, and SVS. Results provided in standard formats like QuPath GeoJSON (polygons), TIFF (heatmaps) and CSV (measurements and statistics).
### Registration and User Access
To start using the Aignostics Platform and its advanced applications, your organization must be registered by our business support team:
1. Access to the Aignostics Platform requires a formal business agreement. Once an agreement is in place between your organization and Aignostics, we proceed with your organization's registration. If your organization does not yet have an account, please contact your account manager or email us at [support@aignostics.com](mailto:support@aignostics.com) to express your interest.
2. To register your organization, we require the name and email address of at least one employee, who will be assigned the Administrator role for your organisation. Your organisation's Administrator can invite and manage additional users.
> [!Important]
> 1. All user accounts must be associated with your organization's official domain. We do not support the registration of private or personal email addresses.
> 2. For security, Two-Factor Authentication (2FA) is mandatory for all user accounts.
> 3. We can integrate with your IDP system (e.g. SAML, OIDC) for user authentication. Please contact us to discuss the integration.
> 4. Registering your organistation typically takes 2 business days depending on the complexity of the signed business agreement and specific requirements.
### Console
The web-based [*Aignostics Console*](https://platform.aignostics.com) is a user-friendly interface that allows you to
manage your organization, applications, quotas, and users registered with the Aignostics Platform.
1. The Console is available to users registered for your organisation to manage their profile and monitor usage of their quota.
2. Administrators of your organization can invite additional users, manage the organisation and user specific quotas and monitor usage.
3. Both roles can trigger application runs.
### Applications
An application is a fully automated advanced machine learning based workflow composed of one or more specific tasks (e.g. Tissue Quality Control, Tissue Segmentation, Cell Detection, Cell Classification and predictive analysis). Each application is designed for a particular analysis purpose (e.g. Tumor Micro Environment analysis or biomarker scoring). For each application we define input requirements, processing tasks and output formats.
As contracted in your business agreement with Aignostics your organisation subscribes to one or more applications. The applications are available for your organization in the Aignostics Platform. You can find the list of available applications in the Console of the Aignostics Platform.
Each application can have multiple versions. Please make sure you read dedicated application documentation to understand its specific constraints regarding acceptable formats, staining method, tissue types and diseases.
Once registered to the Platform, your organization will automatically gain access to the "Test Application". This application can be used to configure the workflow and to make sure that the integration works correctly.
### Application run
To trigger the application run, users can use the Aignostics Launchpad, Aignostics CLI, Example Notebooks, our Client Library, or directly call the REST API. The platform expects the user payload, containing the metadata and the signed URLs to the whole slide images (WSIs). The detailed requirements of the payload depend on the application and are described in the documentation, and accessible via the Info button in the Launchpad, as well as via the CLI and `/v1/applications` endpoint in the API.
When the application run is created, it can be in one of the following states:
1. **received**: the application run received from the client
2. **scheduled**: the application run request is valid and is scheduled for execution
3. **running**: the application run execution started
4. **completed**: the application run execution is done and all outputs are available for download
5. **completed**: the application run execution is done, but some items end up in the failed state
6. **rejected**: the application run request is rejected before it is scheduled
7. **cancelled by the system**: the application run failed during the execution with the number of errors higher than the threshold
9. **cancelled by the user**: the application run is cancelled by the user before it is finished
The status and operations of an application run are private to the user who triggered the run.
### Results
When the processing of whole slide image is successfully completed, the resulting outputs become available for download. To assess specifics of application outputs please consult our application specific documentation, which you can find in the **Console**. Please note that you access to documentation is restricted to those applications your organisation subscribed to.
Application run outputs are automatically deleted 30 days after the application run has completed. However, the owner of the application run (the user who initiated it) can use the API to manually delete outputs earlier, once the run has reached a final state - completed, cancelled by the system or cancelled by the user. The Launchpad and CLI provide enable to delete results with one click resp. command.
### Quotas
Every organization has a limit on how many WSIs it can process in a calendar month. The following quotas exist:
1. **Per organization**: as defined in your business agreement with Aignostics
2. **Per user**: defined by your organization Admin
When the per month quota is reached, an application run request is denied.
Other limitations may apply to your organization:
1. Allowed number of users an organization can register
2. Allowed number of images user can submit per application run
3. Allowed number of parallel application runs for the whole organization
Additionally, we allow organization Admin to define following limitations for its users:
1. Maximum number of images the user can process per calendar month.
2. Maximum number of parallel application runs for a given user
Visit the [Console](https://platform.aignostics.com) to check your current quota and usage. The Console provides a clear overview of the number of images processed by your organization and by each user, as well as the remaining quota for the current month.
### API
The **Aignostics Platform API** is a RESTful web service that allows you to interact with the platform programmatically. It provides endpoints for submitting whole slide images (WSIs) for analysis, checking the status of application runs, and retrieving results.
You can interact with the API using the Python client, which is a wrapper around the RESTful API. The Python client simplifies the process of making requests to the API and handling responses. It also provides convenient methods for uploading WSIs, checking application run status, and downloading results.
For integration with programming languages other than Python, you can use the RESTful API directly. The API is designed to be language-agnostic, meaning you can use any programming language that supports HTTP requests to interact with it. This includes languages like Java, Kotlin, C#, Ruby, and Typescript.
### Cost
Every WSI processed by the Platform generates a cost. Usage of the "Test Application" is free of charge for any registered user. The cost for other applications is defined in your business agreement with Aignostics. The cost is calculated based on the number of slides processed. When an application run is cancelled, either by the system or by the user, only processed images incur a cost.
**[Read the API reference documentation](https://aignostics.readthedocs.io/en/latest/api_reference_v1.html)** or use our **[Interactive API Explorer](https://platform.aignostics.com/explore-api)** to dive into details of all operations and parameters.
## Further Reading
1. Inspect our
[security policy](https://aignostics.readthedocs.io/en/latest/security.html)
with detailed documentation of checks, tools and principles.
1. Inspect how we achieve
[operational excellence](https://aignostics.readthedocs.io/en/latest/operational_excellence.html)
with information on our modern toolchain and software architecture.
2. Check out the
[CLI reference](https://aignostics.readthedocs.io/en/latest/cli_reference.html)
with detailed documentation of all CLI commands and options.
3. Check out the
[library reference](https://aignostics.readthedocs.io/en/latest/lib_reference.html)
with detailed documentation of public classes and functions.
4. Check out the
[API reference](https://aignostics.readthedocs.io/en/latest/api_reference_v1.html)
with detailed documentation of all API operations and parameters. See as well
the OpenAPI Specification in [JSON](https://github.com/aignostics/python-sdk/blob/main/docs/source/_static/openapi_v1.json) and [YAML](https://github.com/aignostics/python-sdk/blob/main/docs/source/_static/openapi_v1.yaml), and the [API Explorer](https://aignostics.readthedocs.io/en/latest/api_explorer_v1.html).
5. Our
[release notes](https://aignostics.readthedocs.io/en/latest/release-notes.html)
provide a complete log of recent improvements and changes.
6. We gratefully acknowledge the numerous
[open source projects](https://aignostics.readthedocs.io/en/latest/attributions.html)
that this project builds upon. Thank you to all these wonderful contributors!
## Glossary
### A
**Administrator Role**
A user role within an organization that has permissions to invite and manage additional users, define user-specific quotas, and monitor organizational usage.
**Aignostics CLI**
Command-Line Interface that allows interaction with the Aignostics Platform directly from terminal or shell scripts, enabling dataset management and application runs.
**Aignostics Client Library**
Python library for seamless integration of the Aignostics Platform with enterprise image management systems and scientific workflows.
**Aignostics Console**
Web-based user interface for managing organizations, applications, quotas, users, and monitoring platform usage.
**Aignostics Launchpad**
Graphical desktop application (available for Mac OS X, Windows, and Linux) that allows users to run computational pathology applications on whole slide images and inspect results with QuPath and Python Notebooks.
**Aignostics Platform**
Comprehensive cloud-based service providing standardized, secure interface for accessing advanced computational pathology applications without requiring specialized expertise or complex infrastructure.
**Aignostics Platform API**
RESTful web service that allows programmatic interaction with the Aignostics Platform, providing endpoints for submitting WSIs, checking application run status, and retrieving results.
**Aignostics Python SDK**
Software Development Kit providing multiple pathways to interact with the Aignostics Platform, including the Launchpad, CLI, Client Library, and example notebooks.
**Application**
Fully automated advanced machine learning workflow composed of specific tasks (e.g., Tissue Quality Control, Tissue Segmentation, Cell Detection, Cell Classification) designed for particular analysis purposes.
**Application Run**
The execution instance of an application on submitted whole slide images, which can be in various states: received, scheduled, running, completed, rejected, cancelled by system, or cancelled by user.
**Application Version**
Specific version of an application with defined input requirements, processing tasks, and output formats. Each application can have multiple versions.
**Atlas H&E-TME**
Advanced computational pathology application for Hematoxylin and Eosin-stained Tumor Microenvironment analysis.
### B
**Base MPP (Microns Per Pixel)**
Metadata parameter specifying the resolution of whole slide images, indicating the physical distance represented by each pixel.
**Business Agreement**
Formal contract between an organization and Aignostics required for platform access, defining quotas, applications, and terms of service.
### C
**Checksum CRC32C**
Cyclic Redundancy Check used to verify data integrity of uploaded whole slide images.
**Client**
The main class in the Aignostics Python SDK used to initialize connections and interact with the platform API.
**Computational Pathology**
Field combining digital pathology with artificial intelligence and machine learning to analyze histopathology slides quantitatively.
**Aignostics Console**
Web-based user interface for managing organizations, applications, quotas, users, and monitoring platform usage.
### D
**DICOM (Digital Imaging and Communications in Medicine)**
Standard format for medical imaging data, supported by the Aignostics Platform for whole slide images.
**Download URL**
Signed URL that allows the Aignostics Platform to securely download image data during processing.
### G
**GeoJSON**
Standard format used by QuPath for representing polygonal annotations and results.
**Google Storage Bucket**
Cloud storage service where users can store whole slide images and generate signed URLs for platform access.
### H
**H&E (Hematoxylin and Eosin)**
Common histological staining method for tissue visualization, used in Atlas H&E-TME application.
**Heatmaps**
Visual representations of analysis results provided in TIFF format showing spatial distribution of measurements.
### I
**IDC (NCI Image Data Commons)**
Public repository of medical imaging data that can be queried and downloaded through the Aignostics CLI.
**IMS (Imaging Management Systems)**
Enterprise systems for managing medical imaging data that can be integrated with the Aignostics Platform.
**Input Artifact**
Data object required for application processing, including the actual data file and associated metadata.
**Input Item**
Individual unit of processing in an application run, containing one or more input artifacts with a unique reference identifier.
**Interactive API Explorer**
Tool for exploring and testing API endpoints and parameters interactively.
### J
**Jupyter**
Popular notebook environment supported by the Aignostics Platform for interactive analysis and visualization.
### L
**LIMS (Laboratory Information Management Systems)**
Laboratory systems that can be integrated with the Aignostics Platform for workflow automation.
### M
**Marimo**
Modern notebook environment supported by the Aignostics Platform as an alternative to Jupyter.
**Metadata**
Descriptive information about whole slide images including dimensions, resolution, tissue type, and disease information required for processing.
**MPP (Microns Per Pixel)**
See Base MPP.
### N
**NCI Image Data Commons (IDC)**
See IDC.
### O
**Operational Excellence**
Aignostics' commitment to high-quality software development practices including A-grade code quality, security scanning, and comprehensive documentation.
### P
**Pyramidal**
Multi-resolution image format that stores the same image at different zoom levels for efficient viewing and processing.
**Python SDK**
Software Development Kit providing multiple pathways to interact with the Aignostics Platform through Python programming language.
### Q
**QuPath**
Open-source software for bioimage analysis that can be launched directly from the Aignostics Launchpad to view results.
**Quota**
Limit on the number of whole slide images an organization or user can process per calendar month, as defined in business agreements.
### R
**Reference**
Unique identifier string for each input item in an application run, used to match results with original inputs.
**Results**
Output data from application processing, including measurements, statistics, heatmaps, and annotations, automatically deleted after 30 days.
**RESTful API**
Architectural style for web services that the Aignostics Platform API follows, enabling language-agnostic integration.
### S
**Self-signed URLs**
Secure URLs with embedded authentication that allow the platform to access user data without exposing credentials.
**SVS**
Aperio ScanScope Virtual Slide format, commonly used for whole slide images and supported by the platform.
### T
**Test Application**
Free application automatically available to all registered organizations for workflow configuration and integration testing.
**TIFF (Tagged Image File Format)**
Standard image format supported for both input whole slide images and output heatmaps.
**Tissue Segmentation**
Computational process of identifying and delineating different tissue regions within histopathology slides.
**TME (Tumor Microenvironment)**
The cellular environment surrounding tumor cells, analyzed by the Atlas H&E-TME application.
**Two-Factor Authentication (2FA)**
Mandatory security requirement for all user accounts on the Aignostics Platform.
### U
**UV**
Modern Python package manager used for dependency management and project setup in the SDK documentation.
**UVX**
Tool for running Python applications directly without explicit installation, used to execute Aignostics CLI commands.
### W
**Whole Slide Image (WSI)**
High-resolution digital image of an entire histopathology slide, the primary input format for computational pathology applications.
**Workflow**
Sequence of automated processing steps within an application that transform input images into analytical results.
Raw data
{
"_id": null,
"home_page": null,
"name": "aignostics",
"maintainer": null,
"docs_url": null,
"requires_python": "<3.14,>=3.11",
"maintainer_email": null,
"keywords": "act, aignostics, atlas, codecov, copier, cyclonedx, detect-secrets, devcontainer, dicom, digital-pathology, docker, git-cliff, image-data-commons, jupyter, machine-learning, marimo, medical-imaging, mypy, nicegui, nox, oe-python-template, openslide, pip-audit, pip-licenses, pre-commit, pydantic, pydicom, pypi, pytest, python, qupath, readthedocs, ruff, sonarcloud, sonarqube, sphinx, typer, uv, whole-slide-imaging",
"author": null,
"author_email": "Helmut Hoffer von Ankershoffen <helmut@aignostics.com>, Andreas Kunft <andreas@aignostics.com>",
"download_url": "https://files.pythonhosted.org/packages/51/f4/d26f0db7a35b3625ad615374663f521b282afa9c18f87fd4d7d900bbf807/aignostics-0.2.196.tar.gz",
"platform": null,
"description": "\n[//]: # (README.md generated from docs/partials/README_*.md)\n\n# \ud83d\udd2cAignostics Python SDK\n\n[](https://github.com/aignostics/python-sdk/blob/main/LICENSE)\n[](https://pypi.org/project/aignostics/)\n[](https://github.com/aignostics/python-sdk/actions/workflows/ci-cd.yml)\n[](https://aignostics.readthedocs.io/en/latest/)\n[](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)\n[](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)\n[](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)\n[](https://codecov.io/gh/aignostics/python-sdk)\n[](https://aignostics.betteruptime.com)\n\n> [!NOTE]\n> The Aignostics Python SDK is in alpha, with [Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) and the [Aignostics Platform](https://platform.aignostics.com) in [early access](https://www.linkedin.com/posts/aignostics_introducing-atlas-he-tme-aignostics-is-activity-7325865745827979265-Sya9?utm_source=share&utm_medium=member_desktop&rcm=ACoAABRmV7cBCGv8eM_ot_kRTrBsb12olQvoLS4). \n> Watch or star this repository to receive updates on new features and improvements of the SDK.\n\n---\n\n\n## Introduction\n\nThe **Aignostics Python SDK** includes multiple pathways to interact with the\n**Aignostics Platform**:\n\n1. Use the **Aignostics Launchpad** to analyze whole slide images with advanced computational pathology applications like\n [Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) directly from your desktop.\n View your results by launching popular tools such as [QuPath](https://qupath.github.io/) and Python Notebooks with one click.\n The app runs on Mac OS X, Windows, and Linux.\n2. Use the **Aignostics Command-line interface (CLI)** to run applications directly from your terminal or shell scripts.\n THe CLI lets you query public datasets from the [NCI Image Data Commons (IDC)](https://portal.imaging.datacommons.cancer.gov/),\n process both public and private whole slide images, and easily download results. The CLI is available for Mac OS X, Windows, and Linux.\n3. Use the included **example notebooks** as starting points to run applications\n directly from your preferred notebook environment. We support Marimo and Jupyter based notebooks environments including Google Collab.\n4. Use the **Aignostics Client Library** to seamlessly integrate the Aignostics Platform with your enterprise image management systems and scientific workflows.\n The client provides a simple way to access the Aignostics Platform API from your Python codebase. We support Python 3.11 and above.\n\n### We take quality and security seriously\n\nWe know you take **quality** and **security** as seriously as we do. That's why\nthe Aignostics Python SDK is built following best practices and with full\ntransparency. This includes (1) making the complete\n[source code of the SDK\navailable on GitHub](https://github.com/aignostics/python-sdk/), maintaining a\n(2)\n[A-grade code quality](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)\nwith [high test coverage](https://app.codecov.io/gh/aignostics/python-sdk) in\nall releases, (3) achieving\n[A-grade security](https://sonarcloud.io/summary/new_code?id=aignostics_python-sdk)\nwith\n[active scanning of dependencies](https://github.com/aignostics/python-sdk/issues/4),\nand (4) providing\n[extensive documentation](hhttps://aignostics.readthedocs.io/en/latest/). Read\nmore about how we achieve\n[operational excellence](https://aignostics.readthedocs.io/en/latest/operational_excellence.html) and\n[security](https://aignostics.readthedocs.io/en/latest/security.html).\n\n## Quick Start\n\n> [!Note]\n> See as follows for a quick start guide to get you up and running with the Aignostics Python SDK as quickly as possible.\n> If you first want to learn bout the basic concepts and components of the Aignostics Platform skip to that section below.\n> The further reading section points you to reference documentation listing all available CLI commands, methods and classes provided by the client library, operations of the API, how we achieve operational excellence, security, and more.\n> If you are not familiar with terminology please check the glossary at the end of this document.\n\n### Launchpad: Run your first computational pathology analysis in 10 minutes from your desktop\n\nThe **Aignostics Launchpad** is a graphical desktop application that allows you to run\napplications on whole slide images (WSIs) from your computer, and inspect results with QuPath and Python Notebooks with one click. It is designed to be user-friendly and intuitive, for use by Research Pathologists and Data Scientists.\n\nThe Launchpad is available for Mac OS X, Windows, and Linux, and can be installed easily:\n\n1. Visit the [Quick Start](https://platform.aignostics.com/getting-started/quick-start)\n page in the Aignostics Console.\n2. Copy the installation script and paste it into your terminal - compatible with MacOS, Windows, and Linux.\n3. Launch the application by running `uvx aignostics launchpad`.\n4. Follow the intuitive graphical interface to analyze public datasets or your own whole slide images\n with [Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) and other computational pathology applications.\n\n> [!Note]\n> The Launchpad features a growing ecosystem of extensions that seamlessly integrate with standard digital pathology tools. To use the Launchpad with all available extensions, run `uvx --from \"aignostics[qupath,marimo]\" aignostics launchpad`. Currently available extensions are:\n>\n> 1. **QuPath extension**: View your application results in [QuPath](https://qupath.github.io/) with a single click. The Launchpad creates QuPath projects on-the-fly.\n> 2. **Marimo extension**: Analyze your application results using [Marimo](https://marimo.io/) notebooks embedded in the Launchpad. You don't have to leave the Launchpad to do real data science.\n\n### CLI: Manage datasets and application runs from your terminal\n\nThe Python SDK includes the **Aignostics CLI**, a Command-Line Interface (CLI) that allows you to\ninteract with the Aignostics Platform directly from your terminal or shell script.\n\nSee as follows for a simple example where we download a sample dataset for the [Atlas\nH&E-TME application](https://www.aignostics.com/products/he-tme-profiling-product), submit an application run, and download the results.\n\n```shell\n# Download a sample dataset from the NCI Image Data Commons (IDC) portal to your current working directory\n# As the dataset id refers to the TCGA LUAD collection, this creates a directory tcga_luad with the DICOM files\nuvx aignostics dataset idc download 1.3.6.1.4.1.5962.99.1.1069745200.1645485340.1637452317744.2.0 data/\n# Prepare the metadata for the application run by creating a metadata.csv, extracting \n# the required metadata from the DICOM files. We furthermore add the required\n# information about the tissue type and disease. TODO (Helmut): Update\nuvx aignostics application run prepare he-tme data/tcga_luad/run.csv data/\n# Edit the metadata.csv to insert the required information about the staining method, tissue type and disease\n# Adapt to your favourite editor\nnano tcga_luad/metadata.csv \n# Upload the metadata.csv and referenced whole slide images to the Aignostics Platform\nuvx aignostics application run upload he-tme data/tcga_luad/run.csv\n# Submit the application run and print tha run id\nuvx aignostics application run submit he-tme data/tcga_luad/run.csv\n# Check the status of the application run you submitted\nuvx aignostics application run list\n# Incrementally download results when they become available\n# Fill in the id from the output in the previous step\nuvx aignostics application run result download APPLICATION_RUN_ID \n```\n\nFor convenience the the `application run execute` command combines preparation, upload, submission and download.\nThe below is equivalent to the above, while adding additionally required metadata using a mapping:\n\n```shell\nuvx aignostics dataset idc download 1.3.6.1.4.1.5962.99.1.1069745200.1645485340.1637452317744.2.0 data/\nuvx aignostics application run execute he-tme data/tcga_luad/run.csv data/ \".*\\.dcm:staining_method=H&E,tissue=LUNG,disease=LUNG_CANCER\"\n```\n\nThe CLI provides extensive help:\n\n```shell\nuvx aignostics --help # list all spaces such as application, dataset, bucket and system, \nuvx aignostics application --help # list subcommands in the application space\nuvx aignostics application run --help # list subcommands in the application run sub-space\nuvx aignostics application run list --help # show help for specific command\nuvx aignostics application run execute --help # show help for another command\n```\n\nCheck out our\n[CLI reference documentation](https://aignostics.readthedocs.io/en/latest/cli_reference.html)\nto learn about all commands and options available.\n\n### Example Notebooks: Interact with the Aignostics Platform from your Python Notebook environment\n\n> [!IMPORTANT]\n> Before you get started, you need to set up your authentication credentials if\n> you did not yet do so! Please visit\n> [your personal dashboard on the Aignostics Platform website](https://platform.aignostics.com/getting-started/quick-start)\n> and follow the steps outlined in the `Use in Python Notebooks` section.\n\nThe Python SDK includes Jupyter and Marimo notebooks to help you get started interacting\nwith the Aignostics Platform in your notebook environment.\n\nThe notebooks showcase the interaction with the Aignostics Platform using our \"Test Application\". To run one them,\nplease follow the steps outlined in the snippet below to clone this repository and start either the\n[Jupyter](https://docs.jupyter.org/en/latest/index.html)\n([examples/notebook.ipynb](https://github.com/aignostics/python-sdk/blob/main/examples/notebook.ipynb))\nor [Marimo](https://marimo.io/)\n([examples/notebook.py](https://github.com/aignostics/python-sdk/blob/main/examples/notebook.py))\nnotebook:\n\n```shell\n# clone the `python-sdk` repository\ngit clone https://github.com/aignostics/python-sdk.git\n# within the cloned repository, install the SDK and all dependencies\nuv sync --all-extras\n# show jupyter example notebook in the browser\nuv run jupyter notebook examples/notebook.ipynb\n# show marimo example notebook in the browser\nuv run marimo edit examples/notebook.py\n```\n\n> [!Note]\n> You can as well run a notebook within the Aignostics Launchpad. To do so, select the\n> Run you want to inspect in the left sidebar, and click the button \"Open in Python Notebook\".\n\n### Client Library: Call the Aignostics Platform API from your Python scripts\n\n> [!IMPORTANT]\\\n> Before you get started, you need to set up your authentication credentials if\n> you did not yet do so! Please visit\n> [your personal dashboard on the Aignostics Platform website](https://platform.aignostics.com/getting-started/quick-start)\n> and follow the steps outlined in the `Enterprise Integration` section.\n\nNext to using the Launchpad, CLI and example notebooks, the Python SDK includes the\n*Aignostics Client Library* for integration with your Python Codebase.\n\nThe following sections outline how to install the Python SDK for this purpose and\ninteract with the Client.\n\n### Installation\n\nThe Aignostics Python SDK is published on the [Python Package Index (PyPI)](https://pypi.org/project/aignostics/),\nis compatible with Python 3.11 and above, and can be installed via via `uv` or `pip`:\n\n**Install with [uv](https://docs.astral.sh/uv/):** If you don't have uv\ninstalled follow [these instructions](https://docs.astral.sh/uv/getting-started/installation/).\n\n```shell\n# Add Aignostics Python SDK as dependency to your project\nuv add aignostics\n```\n\n**Install with [pip](https://pip.pypa.io/en/stable/)**\n\n```shell\n# Add Python SDK as dependency to your project\npip install aignostics\n```\n\n#### Usage\n\nThe following snippet shows how to use the Client to submit an application\nrun:\n\n```python\nfrom aignostics import platform\n\n# initialize the client\nclient = platform.Client()\n# submit an application run\napplication_run = client.runs.submit(\n application_id=\"test-app\",\n items=[\n platform.InputItem(\n external_id=\"slide-1\",\n input_artifacts=[\n platform.InputArtifact(\n name=\"whole_slide_image\",\n download_url=\"<a signed url to download the data>\",\n metadata={\n \"checksum_base64_crc32c\": \"AAAAAA==\",\n \"resolution_mpp\": 0.25,\n \"width_px\": 1000,\n \"height_px\": 1000,\n },\n )\n ],\n ),\n ],\n)\n# wait for the results and download incrementally as they become available\napplication_run.download_to_folder(\"path/to/download/folder\")\n```\n\nPlease look at the notebooks in the `example` folder for a more detailed example\nand read the\n[client reference documentation](https://aignostics.readthedocs.io/en/latest/lib_reference.html)\nto learn about all classes and methods.\n\n##### Defining the input for an application run\n\nWhen creating an application run, you need to specify the `application_id` and optionally the\n`application_version` (version number) of the application you want to run. If you omit the version,\nthe latest version will be used automatically. Additionally, you need to define the input items you\nwant to process in the run. The input items are defined as follows:\n\n```python\nplatform.InputItem(\n external_id=\"1\",\n input_artifacts=[\n platform.InputArtifact(\n name=\"whole_slide_image\", # defined by the application version's input artifact schema\n download_url=\"<a signed url to download the data>\",\n metadata={ # defined by the application version's input artifact schema\n \"checksum_base64_crc32c\": \"N+LWCg==\",\n \"resolution_mpp\": 0.46499982,\n \"width_px\": 3728,\n \"height_px\": 3640,\n },\n )\n ],\n),\n```\n\nFor each item you want to process, you need to provide a unique `reference`\nstring. This is used to identify the item in the results later on. The\n`input_artifacts` field is a list of `InputArtifact` objects, which defines what\ndata & metadata you need to provide for each item. The required artifacts depend\non the application version you want to run - in the case of test application,\nthere is only one artifact required, which is the image to process on. The\nartifact name is defined as `whole_slide_image` for this application.\n\nThe `download_url` is a signed URL that allows the Aignostics Platform to\ndownload the image data later during processing.\n\n#### Self-signed URLs for large files\n\nTo make the whole slide images you want to process available to the Aignostics Platform, you\nneed to provide a signed URL that allows the platform to download the data.\nSelf-signed URLs for files in google storage buckets can be generated using the\n`generate_signed_url`\n([code](https://github.com/aignostics/python-sdk/blob/407e74f7ae89289b70efd86cbda59ec7414050d5/src/aignostics/client/utils.py#L85)).\n\n**We expect that you provide the\n[required credentials](https://cloud.google.com/docs/authentication/application-default-credentials)\nfor the Google Storage Bucket**\n\n\n## Platform\n\n### Overview\n\nThe **Aignostics Platform** is a comprehensive cloud-based service that allows organizations to leverage advanced computational pathology applications without the need for specialized expertise or complex infrastructure. Via its API it provides a standardized, secure interface for accessing Aignostics' portfolio of advanced computational pathology applications. These applications perform machine learning based tissue and cell analysis on histopathology slides, delivering quantitative measurements, visual representations, and detailed statistical data.\n\n\n\n### Key Features\nAignostics Platform offers key features designed to maximize value for its users:\n\n1. **Run Aignostics applications:** Run Aignostics advanced computational pathology applications like [Atlas H&E-TME](https://www.aignostics.com/products/he-tme-profiling-product) on your whole slide images (WSIs) and receive results in a easy to inspect formats.\n2. **Multiple Access Points:** Interact with the platform via various pathways, from **Aignostics Launchpad** (desktop application for MacOS, Windows and Linux), **Aignostics CLI** (command-line interface for your terminal or shell scripts), **Example Notebooks** (we support Jupyter and Marimo), **Aignostics Client Library** (for integration with your Python codebase), or directly through the **API of the Aignostics Platform** (for integration with any programming language). Contact your business partner at Aignostics if you are interested to discuss a direct integration with your Imaging Management Systems (IMS) and Laboratory Information Management Systems (LIMS).\n3. **Secure Data Handling:** Maintain control of your slide data through secure self-signed URLs. Results are automatically deleted after 30 days, and can be deleted earlier by the user.\n4. **High-throughput processing with incremental results delivery:** Submit up to 500 whole slide images (WSI) in one batch request. Access results for individual slides as they completed processing, without having to wait for the entire batch to finish.\n5. **Standard formats:** Support for commonly used image formats in digital pathology such as pyramidal DICOM, TIFF, and SVS. Results provided in standard formats like QuPath GeoJSON (polygons), TIFF (heatmaps) and CSV (measurements and statistics).\n\n### Registration and User Access\n\nTo start using the Aignostics Platform and its advanced applications, your organization must be registered by our business support team:\n\n1. Access to the Aignostics Platform requires a formal business agreement. Once an agreement is in place between your organization and Aignostics, we proceed with your organization's registration. If your organization does not yet have an account, please contact your account manager or email us at [support@aignostics.com](mailto:support@aignostics.com) to express your interest.\n2. To register your organization, we require the name and email address of at least one employee, who will be assigned the Administrator role for your organisation. Your organisation's Administrator can invite and manage additional users. \n\n> [!Important]\n> 1. All user accounts must be associated with your organization's official domain. We do not support the registration of private or personal email addresses.\n> 2. For security, Two-Factor Authentication (2FA) is mandatory for all user accounts.\n> 3. We can integrate with your IDP system (e.g. SAML, OIDC) for user authentication. Please contact us to discuss the integration.\n> 4. Registering your organistation typically takes 2 business days depending on the complexity of the signed business agreement and specific requirements.\n\n### Console\n\nThe web-based [*Aignostics Console*](https://platform.aignostics.com) is a user-friendly interface that allows you to \nmanage your organization, applications, quotas, and users registered with the Aignostics Platform.\n\n1. The Console is available to users registered for your organisation to manage their profile and monitor usage of their quota.\n2. Administrators of your organization can invite additional users, manage the organisation and user specific quotas and monitor usage.\n3. Both roles can trigger application runs.\n\n### Applications\nAn application is a fully automated advanced machine learning based workflow composed of one or more specific tasks (e.g. Tissue Quality Control, Tissue Segmentation, Cell Detection, Cell Classification and predictive analysis). Each application is designed for a particular analysis purpose (e.g. Tumor Micro Environment analysis or biomarker scoring). For each application we define input requirements, processing tasks and output formats.\n\nAs contracted in your business agreement with Aignostics your organisation subscribes to one or more applications. The applications are available for your organization in the Aignostics Platform. You can find the list of available applications in the Console of the Aignostics Platform.\n\nEach application can have multiple versions. Please make sure you read dedicated application documentation to understand its specific constraints regarding acceptable formats, staining method, tissue types and diseases.\n\nOnce registered to the Platform, your organization will automatically gain access to the \"Test Application\". This application can be used to configure the workflow and to make sure that the integration works correctly.\n\n\n### Application run\n\nTo trigger the application run, users can use the Aignostics Launchpad, Aignostics CLI, Example Notebooks, our Client Library, or directly call the REST API. The platform expects the user payload, containing the metadata and the signed URLs to the whole slide images (WSIs). The detailed requirements of the payload depend on the application and are described in the documentation, and accessible via the Info button in the Launchpad, as well as via the CLI and `/v1/applications` endpoint in the API.\n\nWhen the application run is created, it can be in one of the following states:\n\n1. **received**: the application run received from the client\n2. **scheduled**: the application run request is valid and is scheduled for execution\n3. **running**: the application run execution started\n4. **completed**: the application run execution is done and all outputs are available for download\n5. **completed**: the application run execution is done, but some items end up in the failed state\n6. **rejected**: the application run request is rejected before it is scheduled\n7. **cancelled by the system**: the application run failed during the execution with the number of errors higher than the threshold\n9. **cancelled by the user**: the application run is cancelled by the user before it is finished\n\nThe status and operations of an application run are private to the user who triggered the run.\n\n### Results\nWhen the processing of whole slide image is successfully completed, the resulting outputs become available for download. To assess specifics of application outputs please consult our application specific documentation, which you can find in the **Console**. Please note that you access to documentation is restricted to those applications your organisation subscribed to.\n\nApplication run outputs are automatically deleted 30 days after the application run has completed. However, the owner of the application run (the user who initiated it) can use the API to manually delete outputs earlier, once the run has reached a final state - completed, cancelled by the system or cancelled by the user. The Launchpad and CLI provide enable to delete results with one click resp. command.\n\n### Quotas\nEvery organization has a limit on how many WSIs it can process in a calendar month. The following quotas exist:\n\n1. **Per organization**: as defined in your business agreement with Aignostics\n2. **Per user**: defined by your organization Admin\n\nWhen the per month quota is reached, an application run request is denied.\n\nOther limitations may apply to your organization:\n\n1. Allowed number of users an organization can register\n2. Allowed number of images user can submit per application run\n3. Allowed number of parallel application runs for the whole organization\n\nAdditionally, we allow organization Admin to define following limitations for its users:\n\n1. Maximum number of images the user can process per calendar month.\n2. Maximum number of parallel application runs for a given user\n\nVisit the [Console](https://platform.aignostics.com) to check your current quota and usage. The Console provides a clear overview of the number of images processed by your organization and by each user, as well as the remaining quota for the current month.\n\n### API\n\nThe **Aignostics Platform API** is a RESTful web service that allows you to interact with the platform programmatically. It provides endpoints for submitting whole slide images (WSIs) for analysis, checking the status of application runs, and retrieving results.\n\nYou can interact with the API using the Python client, which is a wrapper around the RESTful API. The Python client simplifies the process of making requests to the API and handling responses. It also provides convenient methods for uploading WSIs, checking application run status, and downloading results.\n\nFor integration with programming languages other than Python, you can use the RESTful API directly. The API is designed to be language-agnostic, meaning you can use any programming language that supports HTTP requests to interact with it. This includes languages like Java, Kotlin, C#, Ruby, and Typescript. \n\n### Cost\n\nEvery WSI processed by the Platform generates a cost. Usage of the \"Test Application\" is free of charge for any registered user. The cost for other applications is defined in your business agreement with Aignostics. The cost is calculated based on the number of slides processed. When an application run is cancelled, either by the system or by the user, only processed images incur a cost.\n\n**[Read the API reference documentation](https://aignostics.readthedocs.io/en/latest/api_reference_v1.html)** or use our **[Interactive API Explorer](https://platform.aignostics.com/explore-api)** to dive into details of all operations and parameters.\n\n\n## Further Reading\n\n1. Inspect our\n [security policy](https://aignostics.readthedocs.io/en/latest/security.html)\n with detailed documentation of checks, tools and principles.\n1. Inspect how we achieve\n [operational excellence](https://aignostics.readthedocs.io/en/latest/operational_excellence.html)\n with information on our modern toolchain and software architecture.\n2. Check out the\n [CLI reference](https://aignostics.readthedocs.io/en/latest/cli_reference.html)\n with detailed documentation of all CLI commands and options.\n3. Check out the\n [library reference](https://aignostics.readthedocs.io/en/latest/lib_reference.html)\n with detailed documentation of public classes and functions.\n4. Check out the\n [API reference](https://aignostics.readthedocs.io/en/latest/api_reference_v1.html)\n with detailed documentation of all API operations and parameters. See as well\n the OpenAPI Specification in [JSON](https://github.com/aignostics/python-sdk/blob/main/docs/source/_static/openapi_v1.json) and [YAML](https://github.com/aignostics/python-sdk/blob/main/docs/source/_static/openapi_v1.yaml), and the [API Explorer](https://aignostics.readthedocs.io/en/latest/api_explorer_v1.html).\n5. Our\n [release notes](https://aignostics.readthedocs.io/en/latest/release-notes.html)\n provide a complete log of recent improvements and changes.\n6. We gratefully acknowledge the numerous\n [open source projects](https://aignostics.readthedocs.io/en/latest/attributions.html)\n that this project builds upon. Thank you to all these wonderful contributors!\n\n\n## Glossary\n\n### A\n\n**Administrator Role** \nA user role within an organization that has permissions to invite and manage additional users, define user-specific quotas, and monitor organizational usage.\n\n**Aignostics CLI** \nCommand-Line Interface that allows interaction with the Aignostics Platform directly from terminal or shell scripts, enabling dataset management and application runs.\n\n**Aignostics Client Library** \nPython library for seamless integration of the Aignostics Platform with enterprise image management systems and scientific workflows.\n\n**Aignostics Console** \nWeb-based user interface for managing organizations, applications, quotas, users, and monitoring platform usage.\n\n**Aignostics Launchpad** \nGraphical desktop application (available for Mac OS X, Windows, and Linux) that allows users to run computational pathology applications on whole slide images and inspect results with QuPath and Python Notebooks.\n\n**Aignostics Platform** \nComprehensive cloud-based service providing standardized, secure interface for accessing advanced computational pathology applications without requiring specialized expertise or complex infrastructure.\n\n**Aignostics Platform API** \nRESTful web service that allows programmatic interaction with the Aignostics Platform, providing endpoints for submitting WSIs, checking application run status, and retrieving results.\n\n**Aignostics Python SDK** \nSoftware Development Kit providing multiple pathways to interact with the Aignostics Platform, including the Launchpad, CLI, Client Library, and example notebooks.\n\n**Application** \nFully automated advanced machine learning workflow composed of specific tasks (e.g., Tissue Quality Control, Tissue Segmentation, Cell Detection, Cell Classification) designed for particular analysis purposes.\n\n**Application Run** \nThe execution instance of an application on submitted whole slide images, which can be in various states: received, scheduled, running, completed, rejected, cancelled by system, or cancelled by user.\n\n**Application Version** \nSpecific version of an application with defined input requirements, processing tasks, and output formats. Each application can have multiple versions.\n\n**Atlas H&E-TME** \nAdvanced computational pathology application for Hematoxylin and Eosin-stained Tumor Microenvironment analysis.\n\n### B\n\n**Base MPP (Microns Per Pixel)** \nMetadata parameter specifying the resolution of whole slide images, indicating the physical distance represented by each pixel.\n\n**Business Agreement** \nFormal contract between an organization and Aignostics required for platform access, defining quotas, applications, and terms of service.\n\n### C\n\n**Checksum CRC32C** \nCyclic Redundancy Check used to verify data integrity of uploaded whole slide images.\n\n**Client** \nThe main class in the Aignostics Python SDK used to initialize connections and interact with the platform API.\n\n**Computational Pathology** \nField combining digital pathology with artificial intelligence and machine learning to analyze histopathology slides quantitatively.\n\n**Aignostics Console** \nWeb-based user interface for managing organizations, applications, quotas, users, and monitoring platform usage.\n\n### D\n\n**DICOM (Digital Imaging and Communications in Medicine)** \nStandard format for medical imaging data, supported by the Aignostics Platform for whole slide images.\n\n**Download URL** \nSigned URL that allows the Aignostics Platform to securely download image data during processing.\n\n### G\n\n**GeoJSON** \nStandard format used by QuPath for representing polygonal annotations and results.\n\n**Google Storage Bucket** \nCloud storage service where users can store whole slide images and generate signed URLs for platform access.\n\n### H\n\n**H&E (Hematoxylin and Eosin)** \nCommon histological staining method for tissue visualization, used in Atlas H&E-TME application.\n\n**Heatmaps** \nVisual representations of analysis results provided in TIFF format showing spatial distribution of measurements.\n\n### I\n\n**IDC (NCI Image Data Commons)** \nPublic repository of medical imaging data that can be queried and downloaded through the Aignostics CLI.\n\n**IMS (Imaging Management Systems)** \nEnterprise systems for managing medical imaging data that can be integrated with the Aignostics Platform.\n\n**Input Artifact** \nData object required for application processing, including the actual data file and associated metadata.\n\n**Input Item** \nIndividual unit of processing in an application run, containing one or more input artifacts with a unique reference identifier.\n\n**Interactive API Explorer** \nTool for exploring and testing API endpoints and parameters interactively.\n\n### J\n\n**Jupyter** \nPopular notebook environment supported by the Aignostics Platform for interactive analysis and visualization.\n\n### L\n\n**LIMS (Laboratory Information Management Systems)** \nLaboratory systems that can be integrated with the Aignostics Platform for workflow automation.\n\n### M\n\n**Marimo** \nModern notebook environment supported by the Aignostics Platform as an alternative to Jupyter.\n\n**Metadata** \nDescriptive information about whole slide images including dimensions, resolution, tissue type, and disease information required for processing.\n\n**MPP (Microns Per Pixel)** \nSee Base MPP.\n\n### N\n\n**NCI Image Data Commons (IDC)** \nSee IDC.\n\n### O\n\n**Operational Excellence** \nAignostics' commitment to high-quality software development practices including A-grade code quality, security scanning, and comprehensive documentation.\n\n### P\n\n**Pyramidal** \nMulti-resolution image format that stores the same image at different zoom levels for efficient viewing and processing.\n\n**Python SDK** \nSoftware Development Kit providing multiple pathways to interact with the Aignostics Platform through Python programming language.\n\n### Q\n\n**QuPath** \nOpen-source software for bioimage analysis that can be launched directly from the Aignostics Launchpad to view results.\n\n**Quota** \nLimit on the number of whole slide images an organization or user can process per calendar month, as defined in business agreements.\n\n### R\n\n**Reference** \nUnique identifier string for each input item in an application run, used to match results with original inputs.\n\n**Results** \nOutput data from application processing, including measurements, statistics, heatmaps, and annotations, automatically deleted after 30 days.\n\n**RESTful API** \nArchitectural style for web services that the Aignostics Platform API follows, enabling language-agnostic integration.\n\n### S\n\n**Self-signed URLs** \nSecure URLs with embedded authentication that allow the platform to access user data without exposing credentials.\n\n**SVS** \nAperio ScanScope Virtual Slide format, commonly used for whole slide images and supported by the platform.\n\n### T\n\n**Test Application** \nFree application automatically available to all registered organizations for workflow configuration and integration testing.\n\n**TIFF (Tagged Image File Format)** \nStandard image format supported for both input whole slide images and output heatmaps.\n\n**Tissue Segmentation** \nComputational process of identifying and delineating different tissue regions within histopathology slides.\n\n**TME (Tumor Microenvironment)** \nThe cellular environment surrounding tumor cells, analyzed by the Atlas H&E-TME application.\n\n**Two-Factor Authentication (2FA)** \nMandatory security requirement for all user accounts on the Aignostics Platform.\n\n### U\n\n**UV** \nModern Python package manager used for dependency management and project setup in the SDK documentation.\n\n**UVX** \nTool for running Python applications directly without explicit installation, used to execute Aignostics CLI commands.\n\n### W\n\n**Whole Slide Image (WSI)** \nHigh-resolution digital image of an entire histopathology slide, the primary input format for computational pathology applications.\n\n**Workflow** \nSequence of automated processing steps within an application that transform input images into analytical results.\n",
"bugtrack_url": null,
"license": "MIT License Copyright (c) [2025] [Aignostics GmbH (support@aignostics.com)] Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the \"Software\"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software. THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.",
"summary": "\ud83d\udd2c Python SDK providing access to the Aignostics Platform. Includes Aignostics Launchpad (Desktop Application), Aignostics CLI (Command-Line Interface), example notebooks, and Aignostics Client Library.",
"version": "0.2.196",
"project_urls": {
"Changelog": "https://github.com/aignostics/python-sdk/releases",
"Documentation": "https://aignostics.readthedocs.io/en/latest/",
"Homepage": "https://aignostics.readthedocs.io/en/latest/",
"Issues": "https://github.com/aignostics/python-sdk/issues",
"Source": "https://github.com/aignostics/python-sdk"
},
"split_keywords": [
"act",
" aignostics",
" atlas",
" codecov",
" copier",
" cyclonedx",
" detect-secrets",
" devcontainer",
" dicom",
" digital-pathology",
" docker",
" git-cliff",
" image-data-commons",
" jupyter",
" machine-learning",
" marimo",
" medical-imaging",
" mypy",
" nicegui",
" nox",
" oe-python-template",
" openslide",
" pip-audit",
" pip-licenses",
" pre-commit",
" pydantic",
" pydicom",
" pypi",
" pytest",
" python",
" qupath",
" readthedocs",
" ruff",
" sonarcloud",
" sonarqube",
" sphinx",
" typer",
" uv",
" whole-slide-imaging"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "ba036f0376d58bd886013056eb45f9bd2537f653c348ad625591a255728b39fd",
"md5": "3595b07daec5f75f23c812f7391676f3",
"sha256": "ac6323abc8311e65d6a554fbd49ee4fd6ee9669a92ebb0a0db4b1e922790e999"
},
"downloads": -1,
"filename": "aignostics-0.2.196-py3-none-any.whl",
"has_sig": false,
"md5_digest": "3595b07daec5f75f23c812f7391676f3",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": "<3.14,>=3.11",
"size": 4058832,
"upload_time": "2025-10-26T23:41:35",
"upload_time_iso_8601": "2025-10-26T23:41:35.491120Z",
"url": "https://files.pythonhosted.org/packages/ba/03/6f0376d58bd886013056eb45f9bd2537f653c348ad625591a255728b39fd/aignostics-0.2.196-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "51f4d26f0db7a35b3625ad615374663f521b282afa9c18f87fd4d7d900bbf807",
"md5": "26822f5ce57dda020c71e9c89b889f83",
"sha256": "869a3449d3339b7178e57b2a84a627c70b321a8fd3f5f0e4334b1653f8bbecda"
},
"downloads": -1,
"filename": "aignostics-0.2.196.tar.gz",
"has_sig": false,
"md5_digest": "26822f5ce57dda020c71e9c89b889f83",
"packagetype": "sdist",
"python_version": "source",
"requires_python": "<3.14,>=3.11",
"size": 3981979,
"upload_time": "2025-10-26T23:41:37",
"upload_time_iso_8601": "2025-10-26T23:41:37.713583Z",
"url": "https://files.pythonhosted.org/packages/51/f4/d26f0db7a35b3625ad615374663f521b282afa9c18f87fd4d7d900bbf807/aignostics-0.2.196.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-10-26 23:41:37",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "aignostics",
"github_project": "python-sdk",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "aignostics"
}
