<img src="https://raw.githubusercontent.com/AMICI-dev/AMICI/main/doc/gfx/banner.png" height="60" align="left" alt="AMICI logo">
## Advanced Multilanguage Interface for CVODES and IDAS
## About
AMICI provides a multi-language (Python, C++, Matlab) interface for the
[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers
[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)
(for ordinary differential equations) and
[IDAS](https://computing.llnl.gov/projects/sundials/idas)
(for algebraic differential equations). AMICI allows the user to read
differential equation models specified as [SBML](http://sbml.org/)
or [PySB](http://pysb.org/)
and automatically compiles such models into Python modules, C++ libraries or
Matlab `.mex` simulation files.
The generated model expressions along with the corresponding sensitivity
equations are transformed into native C++ code which allows for a significantly
faster simulation.
**NOTE: The MATLAB interface is no longer supported and will be removed soon.**
Beyond forward integration, the compiled simulation file also allows for
forward sensitivity analysis, steady state sensitivity analysis and
adjoint sensitivity analysis for likelihood-based output functions.
The interface was designed to provide routines for efficient gradient
computation in parameter estimation of biochemical reaction models, but
it is also applicable to a wider range of differential equation
constrained optimization problems.
## Current build status
<a href="https://badge.fury.io/py/amici">
<img src="https://badge.fury.io/py/amici.svg" alt="PyPI version"></a>
<a href="https://github.com/AMICI-dev/AMICI/actions/workflows/test_pypi.yml">
<img src="https://github.com/AMICI-dev/AMICI/actions/workflows/test_pypi.yml/badge.svg" alt="PyPI installation"></a>
<a href="https://codecov.io/gh/AMICI-dev/AMICI">
<img src="https://codecov.io/gh/AMICI-dev/AMICI/branch/main/graph/badge.svg" alt="Code coverage"></a>
<a href="https://sonarcloud.io/dashboard?id=ICB-DCM_AMICI&branch=main">
<img src="https://sonarcloud.io/api/project_badges/measure?project=ICB-DCM_AMICI&metric=sqale_index" alt="SonarCloud technical debt"></a>
<a href="https://zenodo.org/badge/latestdoi/43677177">
<img src="https://zenodo.org/badge/43677177.svg" alt="Zenodo DOI"></a>
<a href="https://amici.readthedocs.io/en/latest/?badge=latest">
<img src="https://readthedocs.org/projects/amici/badge/?version=latest" alt="ReadTheDocs status"></a>
<a href="https://bestpractices.coreinfrastructure.org/projects/3780">
<img src="https://bestpractices.coreinfrastructure.org/projects/3780/badge" alt="coreinfrastructure bestpractices badge"></a>
## Features
* SBML import
* PySB import
* Generation of C++ code for model simulation and sensitivity
computation
* Access to and high customizability of CVODES and IDAS solver
* Python, C++, Matlab interface
* Sensitivity analysis
* forward
* steady state
* adjoint
* first- and second-order
* Pre-equilibration and pre-simulation conditions
* Support for
[discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)
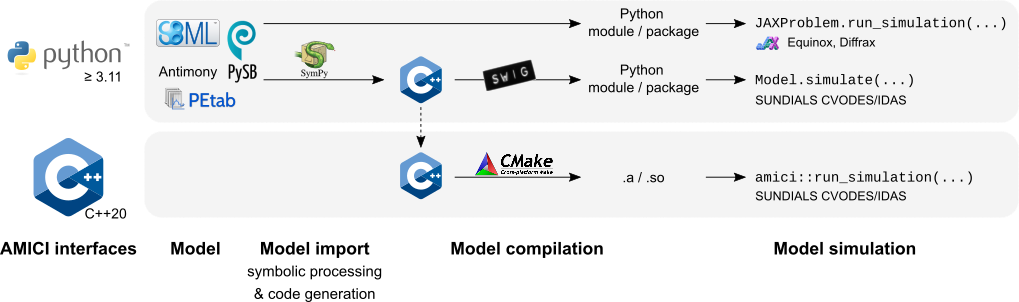
## Interfaces & workflow
The AMICI workflow starts with importing a model from either
[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),
or a Matlab definition of the model (Matlab-only). From this input,
all equations for model simulation
are derived symbolically and C++ code is generated. This code is then
compiled into a C++ library, a Python module, or a Matlab `.mex` file and
is then used for model simulation.
## Getting started
The AMICI source code is available at https://github.com/AMICI-dev/AMICI/.
To install AMICI, first read the installation instructions for
[Python](https://amici.readthedocs.io/en/latest/python_installation.html),
[C++](https://amici.readthedocs.io/en/latest/cpp_installation.html) or
[Matlab](https://amici.readthedocs.io/en/latest/matlab_installation.html).
There are also instructions for using AMICI inside
[containers](https://github.com/AMICI-dev/AMICI/tree/main/container).
To get you started with Python-AMICI, the best way might be checking out this
[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/main/doc/examples/getting_started/GettingStarted.ipynb)
[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/main?labpath=doc%2Fexamples%2Fgetting_started%2FGettingStarted.ipynb).
To get started with Matlab-AMICI, various examples are available
in [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/main/matlab/examples).
Comprehensive documentation is available at
[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).
Any [contributions](https://amici.readthedocs.io/en/latest/CONTRIBUTING.html)
to AMICI are welcome (code, bug reports, suggestions for improvements, ...).
## Getting help
In case of questions or problems with using AMICI, feel free to post an
[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to
get back to you quickly.
## Projects using AMICI
There are several tools for parameter estimation offering good integration
with AMICI:
* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for
optimization, sampling and uncertainty analysis
* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for
parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential
Monte Carlo)
* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter
estimation of ODE models offering distributed memory parallelism with focus
on problems with many simulation conditions.
## Publications
**Citeable DOI for the latest AMICI release:**
[](https://zenodo.org/badge/latestdoi/43677177)
There is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).
If you used AMICI in your work, we are happy to include
your project, please let us know via a GitHub issue.
When using AMICI in your project, please cite:
* Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.
AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,
[DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).
```bibtex
@article{frohlich2020amici,
title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},
author={Fr{\"o}hlich, Fabian and Weindl, Daniel and Sch{\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},
journal = {Bioinformatics},
year = {2021},
month = {04},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btab227},
note = {btab227},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},
}
```
When presenting work that employs AMICI, feel free to use one of the icons in
[doc/gfx/](https://github.com/AMICI-dev/AMICI/tree/main/doc/gfx),
which are available under a
[CC0](https://github.com/AMICI-dev/AMICI/tree/main/doc/gfx/LICENSE.md)
license:
<p align="center">
<img src="https://raw.githubusercontent.com/AMICI-dev/AMICI/main/doc/gfx/logo_text.png" height="75" alt="AMICI Logo">
</p>
Raw data
{
"_id": null,
"home_page": null,
"name": "amici",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.11",
"maintainer_email": "Fabian Froehlich <froehlichfab@gmail.com>, Daniel Weindl <sci@danielweindl.de>",
"keywords": "differential equations, simulation, ode, cvodes, systems biology, sensitivity analysis, sbml, pysb, petab",
"author": "Jan Hasenauer, AMICI contributors",
"author_email": "Fabian Froehlich <froehlichfab@gmail.com>, Daniel Weindl <sci@danielweindl.de>",
"download_url": "https://files.pythonhosted.org/packages/40/26/f83bfbd35bcff63644e5225842fee68da91fcdfc31dda94406cb5d88c325/amici-0.34.1.tar.gz",
"platform": null,
"description": "<img src=\"https://raw.githubusercontent.com/AMICI-dev/AMICI/main/doc/gfx/banner.png\" height=\"60\" align=\"left\" alt=\"AMICI logo\">\n\n## Advanced Multilanguage Interface for CVODES and IDAS\n\n## About\n\nAMICI provides a multi-language (Python, C++, Matlab) interface for the\n[SUNDIALS](https://computing.llnl.gov/projects/sundials/) solvers\n[CVODES](https://computing.llnl.gov/projects/sundials/cvodes)\n(for ordinary differential equations) and\n[IDAS](https://computing.llnl.gov/projects/sundials/idas)\n(for algebraic differential equations). AMICI allows the user to read\ndifferential equation models specified as [SBML](http://sbml.org/)\nor [PySB](http://pysb.org/)\nand automatically compiles such models into Python modules, C++ libraries or\nMatlab `.mex` simulation files.\nThe generated model expressions along with the corresponding sensitivity\nequations are transformed into native C++ code which allows for a significantly\nfaster simulation.\n\n**NOTE: The MATLAB interface is no longer supported and will be removed soon.**\n\nBeyond forward integration, the compiled simulation file also allows for\nforward sensitivity analysis, steady state sensitivity analysis and\nadjoint sensitivity analysis for likelihood-based output functions.\n\nThe interface was designed to provide routines for efficient gradient\ncomputation in parameter estimation of biochemical reaction models, but\nit is also applicable to a wider range of differential equation\nconstrained optimization problems.\n\n## Current build status\n\n<a href=\"https://badge.fury.io/py/amici\">\n <img src=\"https://badge.fury.io/py/amici.svg\" alt=\"PyPI version\"></a>\n<a href=\"https://github.com/AMICI-dev/AMICI/actions/workflows/test_pypi.yml\">\n <img src=\"https://github.com/AMICI-dev/AMICI/actions/workflows/test_pypi.yml/badge.svg\" alt=\"PyPI installation\"></a>\n<a href=\"https://codecov.io/gh/AMICI-dev/AMICI\">\n <img src=\"https://codecov.io/gh/AMICI-dev/AMICI/branch/main/graph/badge.svg\" alt=\"Code coverage\"></a>\n<a href=\"https://sonarcloud.io/dashboard?id=ICB-DCM_AMICI&branch=main\">\n <img src=\"https://sonarcloud.io/api/project_badges/measure?project=ICB-DCM_AMICI&metric=sqale_index\" alt=\"SonarCloud technical debt\"></a>\n<a href=\"https://zenodo.org/badge/latestdoi/43677177\">\n <img src=\"https://zenodo.org/badge/43677177.svg\" alt=\"Zenodo DOI\"></a>\n<a href=\"https://amici.readthedocs.io/en/latest/?badge=latest\">\n <img src=\"https://readthedocs.org/projects/amici/badge/?version=latest\" alt=\"ReadTheDocs status\"></a>\n<a href=\"https://bestpractices.coreinfrastructure.org/projects/3780\">\n <img src=\"https://bestpractices.coreinfrastructure.org/projects/3780/badge\" alt=\"coreinfrastructure bestpractices badge\"></a>\n\n## Features\n\n* SBML import\n* PySB import\n* Generation of C++ code for model simulation and sensitivity\n computation\n* Access to and high customizability of CVODES and IDAS solver\n* Python, C++, Matlab interface\n* Sensitivity analysis\n * forward\n * steady state\n * adjoint\n * first- and second-order\n* Pre-equilibration and pre-simulation conditions\n* Support for\n [discrete events and logical operations](https://academic.oup.com/bioinformatics/article/33/7/1049/2769435)\n\n## Interfaces & workflow\n\nThe AMICI workflow starts with importing a model from either\n[SBML](http://sbml.org/) (Matlab, Python), [PySB](http://pysb.org/) (Python),\nor a Matlab definition of the model (Matlab-only). From this input,\nall equations for model simulation\nare derived symbolically and C++ code is generated. This code is then\ncompiled into a C++ library, a Python module, or a Matlab `.mex` file and\nis then used for model simulation.\n\n\n\n## Getting started\n\nThe AMICI source code is available at https://github.com/AMICI-dev/AMICI/.\nTo install AMICI, first read the installation instructions for\n[Python](https://amici.readthedocs.io/en/latest/python_installation.html),\n[C++](https://amici.readthedocs.io/en/latest/cpp_installation.html) or\n[Matlab](https://amici.readthedocs.io/en/latest/matlab_installation.html).\nThere are also instructions for using AMICI inside\n[containers](https://github.com/AMICI-dev/AMICI/tree/main/container).\n\nTo get you started with Python-AMICI, the best way might be checking out this\n[Jupyter notebook](https://github.com/AMICI-dev/AMICI/blob/main/doc/examples/getting_started/GettingStarted.ipynb)\n[](https://mybinder.org/v2/gh/AMICI-dev/AMICI/main?labpath=doc%2Fexamples%2Fgetting_started%2FGettingStarted.ipynb).\n\nTo get started with Matlab-AMICI, various examples are available\nin [matlab/examples/](https://github.com/AMICI-dev/AMICI/tree/main/matlab/examples).\n\nComprehensive documentation is available at\n[https://amici.readthedocs.io/en/latest/](https://amici.readthedocs.io/en/latest/).\n\nAny [contributions](https://amici.readthedocs.io/en/latest/CONTRIBUTING.html)\nto AMICI are welcome (code, bug reports, suggestions for improvements, ...).\n\n\n## Getting help\n\nIn case of questions or problems with using AMICI, feel free to post an\n[issue](https://github.com/AMICI-dev/AMICI/issues) on GitHub. We are trying to\nget back to you quickly.\n\n## Projects using AMICI\n\nThere are several tools for parameter estimation offering good integration\nwith AMICI:\n\n* [pyPESTO](https://github.com/ICB-DCM/pyPESTO): Python library for\n optimization, sampling and uncertainty analysis\n* [pyABC](https://github.com/ICB-DCM/pyABC): Python library for\n parallel and scalable ABC-SMC (Approximate Bayesian Computation - Sequential\n Monte Carlo)\n* [parPE](https://github.com/ICB-DCM/parPE): C++ library for parameter\n estimation of ODE models offering distributed memory parallelism with focus\n on problems with many simulation conditions.\n\n## Publications\n\n**Citeable DOI for the latest AMICI release:**\n[](https://zenodo.org/badge/latestdoi/43677177)\n\nThere is a list of [publications using AMICI](https://amici.readthedocs.io/en/latest/references.html).\nIf you used AMICI in your work, we are happy to include\nyour project, please let us know via a GitHub issue.\n\nWhen using AMICI in your project, please cite:\n\n* Fr\u00f6hlich, F., Weindl, D., Sch\u00e4lte, Y., Pathirana, D., Paszkowski, \u0141., Lines, G.T., Stapor, P. and Hasenauer, J., 2021.\n AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227,\n [DOI:10.1093/bioinformatics/btab227](https://doi.org/10.1093/bioinformatics/btab227).\n\n ```bibtex\n @article{frohlich2020amici,\n title={AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models},\n author={Fr{\\\"o}hlich, Fabian and Weindl, Daniel and Sch{\\\"a}lte, Yannik and Pathirana, Dilan and Paszkowski, {\\L}ukasz and Lines, Glenn Terje and Stapor, Paul and Hasenauer, Jan},\n journal = {Bioinformatics},\n year = {2021},\n month = {04},\n issn = {1367-4803},\n doi = {10.1093/bioinformatics/btab227},\n note = {btab227},\n eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btab227/36866220/btab227.pdf},\n }\n ```\n\nWhen presenting work that employs AMICI, feel free to use one of the icons in\n[doc/gfx/](https://github.com/AMICI-dev/AMICI/tree/main/doc/gfx),\nwhich are available under a\n[CC0](https://github.com/AMICI-dev/AMICI/tree/main/doc/gfx/LICENSE.md)\nlicense:\n\n<p align=\"center\">\n <img src=\"https://raw.githubusercontent.com/AMICI-dev/AMICI/main/doc/gfx/logo_text.png\" height=\"75\" alt=\"AMICI Logo\">\n</p>\n",
"bugtrack_url": null,
"license": null,
"summary": "Advanced multi-language Interface to CVODES and IDAS",
"version": "0.34.1",
"project_urls": {
"Bug Tracker": "https://github.com/AMICI-dev/AMICI/issues",
"Documentation": "https://amici.readthedocs.io/en/latest/",
"Homepage": "https://github.com/AMICI-dev/AMICI",
"Repository": "https://github.com/AMICI-dev/AMICI.git"
},
"split_keywords": [
"differential equations",
" simulation",
" ode",
" cvodes",
" systems biology",
" sensitivity analysis",
" sbml",
" pysb",
" petab"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "4026f83bfbd35bcff63644e5225842fee68da91fcdfc31dda94406cb5d88c325",
"md5": "3e4245b92613ccc949b0fde416cc0403",
"sha256": "edb2fa77d109f128921e111ce303eec0842723138c1d3863b0c5cbdd733039e1"
},
"downloads": -1,
"filename": "amici-0.34.1.tar.gz",
"has_sig": false,
"md5_digest": "3e4245b92613ccc949b0fde416cc0403",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.11",
"size": 1221947,
"upload_time": "2025-08-27T08:37:02",
"upload_time_iso_8601": "2025-08-27T08:37:02.560600Z",
"url": "https://files.pythonhosted.org/packages/40/26/f83bfbd35bcff63644e5225842fee68da91fcdfc31dda94406cb5d88c325/amici-0.34.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-08-27 08:37:02",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "AMICI-dev",
"github_project": "AMICI",
"travis_ci": false,
"coveralls": true,
"github_actions": true,
"tox": true,
"lcname": "amici"
}

