| Name | biapy JSON |
| Version |
3.6.4
 JSON
JSON |
| download |
| home_page | None |
| Summary | BiaPy: Bioimage analysis pipelines in Python |
| upload_time | 2025-08-15 07:17:27 |
| maintainer | None |
| docs_url | None |
| author | None |
| requires_python | >=3.10 |
| license | MIT License
Copyright (c) 2022 Daniel Franco-Barranco
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
|
| keywords |
feed
reader
tutorial
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|

# BiaPy: Accessible deep learning on bioimages
<p align="left">
<a href="https://www.python.org/">
<img src="https://img.shields.io/badge/Python-3.10-yellow.svg" /></a>
<a href= "https://pytorch.org/">
<img src="https://img.shields.io/badge/PyTorch-2.4-orange.svg" /></a>
<a href= "https://pypi.org/project/biapy/">
<img src="https://img.shields.io/pypi/v/biapy.svg?color=green" /></a>
<a href= "https://anaconda.org/conda-forge/biapy/">
<img src="https://anaconda.org/conda-forge/biapy/badges/version.svg" /></a>
<a href= "https://github.com/BiaPyX/BiaPy/blob/master/LICENSE">
<img src="https://img.shields.io/badge/License-MIT-blue.svg" /></a>
<a href= "https://biapy.readthedocs.io/en/latest/">
<img src="https://img.shields.io/badge/Doc-Latest-2BAF2B.svg" /></a>
<a href= "https://www.nature.com/articles/s41592-025-02699-y">
<img src="https://img.shields.io/badge/Nature_Methods-Paper-bd2635.svg" /></a>
<a href= "https://ieeexplore.ieee.org/abstract/document/10230593">
<img src="https://img.shields.io/badge/IEEE-Paper-00629B.svg" /></a>
</p>
[](https://github.com/BiaPyX/BiaPy/actions/workflows/check_code_consistency.yml)
[](https://github.com/BiaPyX/BiaPy/actions/workflows/upload_biapy_to_pypi.yml)
[](https://github.com/BiaPyX/BiaPy/actions/workflows/create_release_container.yml)
🔥**NEWS**🔥: BiaPy's paper is finally out in **Nature Methods**! Check it out at: [https://www.nature.com/articles/s41592-025-02699-y](https://www.nature.com/articles/s41592-025-02699-y)
For those who have no access to Nature Methods, here is the **preprint** at bioRxiv: [https://www.biorxiv.org/content/10.1101/2024.02.03.576026v3](https://www.biorxiv.org/content/10.1101/2024.02.03.576026v3)
[BiaPy](https://biapyx.github.io) is an open source library and application that streamlines the use of common deep-learning workflows for a large variety of bioimage analysis tasks, including 2D and 3D [semantic segmentation](https://biapy.readthedocs.io/en/latest/workflows/semantic_segmentation.html), [instance segmentation](https://biapy.readthedocs.io/en/latest/workflows/instance_segmentation.html), [object detection](https://biapy.readthedocs.io/en/latest/workflows/detection.html), [image denoising](https://biapy.readthedocs.io/en/latest/workflows/denoising.html), [single image super-resolution](https://biapy.readthedocs.io/en/latest/workflows/super_resolution.html), [self-supervised learning](https://biapy.readthedocs.io/en/latest/workflows/self_supervision.html), [image classification](https://biapy.readthedocs.io/en/latest/workflows/classification.html) and [image to image translation](https://biapy.readthedocs.io/en/latest/workflows/image_to_image.html).
BiaPy is a versatile platform designed to accommodate both proficient computer scientists and users less experienced in programming. It offers diverse and user-friendly access points to our workflows.
This repository is actively under development by the [BiaPy Team](https://biapyx.github.io/about/), a group of contributors from different research institutions.

## Description videos
Find a comprehensive overview of BiaPy and its functionality in the following videos:
| [](https://www.youtube.com/watch?v=Gnm-VsZQ6Cc "BiaPy: a ready-to-use library for Bioimage Analysis Pipelines") <br> <span style="font-weight:normal">BiaPy history and GUI demo at [RTmfm](https://rtmfm.cnrs.fr/en/) by Ignacio Arganda-Carreras and Daniel Franco-Barranco.</span> |
|-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
| [](https://youtu.be/6eYtX-ySpc0 "BiaPy: a ready-to-use library for Bioimage Analysis Pipelines") <br> BiaPy presentation at [Virtual Pub of Euro-BioImaging](https://www.eurobioimaging.eu/) by Ignacio Arganda-Carreras. | | | | |
## User interface
You can also use BiaPy through our graphical user interface (GUI).
## Download BiaPy GUI for you OS
- [Windows 64-bit](https://drive.google.com/uc?export=download&id=1iV0wzdFhpCpBCBgsameGyT3iFyQ6av5o)
- [Linux 64-bit](https://drive.google.com/uc?export=download&id=13jllkLTR6S3yVZLRdMwhWUu7lq3HyJsD)
- [macOS 64-bit](https://drive.google.com/uc?export=download&id=1fIpj9A8SWIN1fABEUAS--DNhOHzqSL7f)
Project's page: [[BiaPy GUI](https://github.com/BiaPyX/BiaPy-GUI)]
## Scientific publications using BiaPy
| | |
|--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|:-----------------------------------------------------------------------------------------------------------------------------------------------------------:|
| ```López-Cano, Daniel, et al. "Characterizing Structure Formation through Instance Segmentation" (2023).``` <br><br> This study presents a machine-learning framework to predict the formation of dark matter haloes from early universe density perturbations. Utilizing two neural networks, it distinguishes particles comprising haloes and groups them by membership. The framework accurately predicts halo masses and shapes, and compares favorably with N-body simulations. The open-source model could enhance analytical methods of structure formation by analyzing initial condition variations. ```BiaPy``` is used in the creation of the watershed approach. <br><br> [[Documentation (not yet)](https://biapy.readthedocs.io/en/latest)] [[Paper](https://arxiv.org/abs/2311.12110)] | 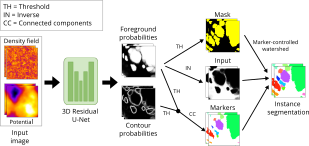 |
| ```Franco-Barranco, Daniel, et al. "Current Progress and Challenges in Large-scale 3D Mitochondria Instance Segmentation." (2023).``` <br><br> This paper reports the results of the MitoEM challenge on 3D instance segmentation of mitochondria in electron microscopy images, held in conjunction with IEEE-ISBI 2021. The paper discusses the top-performing methods, addresses ground truth errors, and proposes a new scoring system to improve segmentation evaluation. Despite progress, challenges remain in segmenting mitochondria with complex shapes, keeping the competition open for further submissions. ```BiaPy``` is used in the creation of the MitoEM challenge baseline (U2D-BC). <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/mitoem.html)] [[Paper](https://ieeexplore.ieee.org/document/10266382)] [[Toolbox](https://github.com/BiaPyX/TIMISE)] | 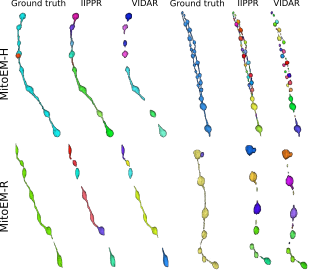 |
| ```Backová, Lenka, et al. "Modeling Wound Healing Using Vector Quantized Variational Autoencoders and Transformers." 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI). IEEE, 2023.``` <br><br> This study focuses on time-lapse sequences of Drosophila embryos healing from laser-incised wounds. The researchers employ a two-stage approach involving a vector quantized variational autoencoder and an autoregressive transformer to model wound healing as a video prediction task. ```BiaPy``` is used in the creation of the wound segmentation masks. <br><br> [[Documentation](https://lenkaback.github.io/wound-healing-modeling/)] [[Paper](https://ieeexplore.ieee.org/document/10230571)] | 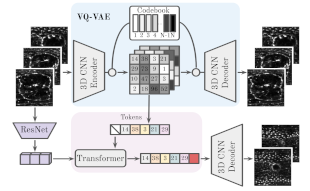 |
| ```Andrés-San Román, Jesús A., et al. "CartoCell, a high-content pipeline for 3D image analysis, unveils cell morphology patterns in epithelia." Cell Reports Methods (2023)``` <br><br> Combining deep learning and 3D imaging is crucial for high-content analysis. CartoCell is introduced, a method that accurately labels 3D epithelial cysts, enabling quantification of cellular features and mapping their distribution. It's adaptable to other epithelial tissues. CartoCell method is created using ```BiaPy```. <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/cartocell.html)] [[Paper](https://www.cell.com/cell-reports-methods/pdf/S2667-2375(23)00249-7.pdf)] | <table> <tr> <td> </td> <td> </td> </tr> </table> |
| ```Franco-Barranco, Daniel, et al. "Deep learning based domain adaptation for mitochondria segmentation on EM volumes." Computer Methods and Programs in Biomedicine 222 (2022): 106949.``` <br><br> This study addresses mitochondria segmentation across different datasets using three unsupervised domain adaptation approaches, including style transfer, self-supervised learning, and multi-task neural networks. To ensure robust generalization, a new training stopping criterion based on source domain morphological priors is proposed. ```BiaPy``` is used for the implementation of the Attention U-Net. <br><br> [[Documentation](https://github.com/danifranco/EM_domain_adaptation)] [[Paper](https://www.sciencedirect.com/science/article/pii/S0169260722003315)] | 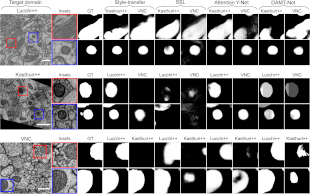 |
| ```Franco-Barranco, Daniel, et al. "Stable deep neural network architectures for mitochondria segmentation on electron microscopy volumes." Neuroinformatics 20.2 (2022): 437-450.``` <br><br> Recent deep learning models have shown impressive performance in mitochondria segmentation, but often lack code and training details, affecting reproducibility. This study follows best practices, comprehensively comparing state-of-the-art architectures and variations of U-Net models for mitochondria segmentation, revealing their impact and stability. The research consistently achieves state-of-the-art results on various datasets, including EPFL Hippocampus, Lucchi++, and Kasthuri++. ```BiaPy``` is used for the implementation of the methods compared in the study. <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/stable.html)] [[Paper](https://link.springer.com/article/10.1007/s12021-021-09556-1)] | 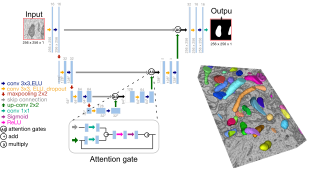 |
| ```Wei, Donglai, et al. "Mitoem dataset: Large-scale 3d mitochondria instance segmentation from em images." International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer International Publishing, 2020.``` <br><br> Existing mitochondria segmentation datasets are small, raising questions about method robustness. The MitoEM dataset introduces larger 3D volumes with diverse mitochondria, challenging existing instance segmentation methods, highlighting the need for improved techniques. ```BiaPy``` is used in the creation of the MitoEM challenge baseline (U2D-BC). <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/mitoem.html)] [[Paper](https://link.springer.com/chapter/10.1007/978-3-030-59722-1_7)] [[Challenge](https://mitoem.grand-challenge.org/)] | 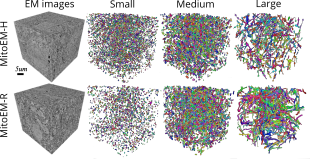 |
## Authors
| Name | Role | Affiliations |
|------------------------------------------------------------------------------------|------------------------------------------|---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
| [Daniel Franco-Barranco](https://BiaPyX.github.io/) | Creator, Implementation, Software design | <ul> <li> Dept. of Computer Science and Artificial Intelligence, University of the Basque Country (UPV/EHU) </li> <li> Donostia International Physics Center (DIPC) </li> </ul> |
| [Lenka Backová](https://www.biofisika.org/en/about/people/lenka-backova) | Implementation | <ul> <li> Biofisika Institute </li> </ul> |
| [Aitor Gonzalez-Marfil](https://dipc.ehu.eus/es/dipc/personas/investigadores-en-formacion/aitor-gonzalez-marfil) | Implementation | <ul> <li> Dept. of Computer Science and Artificial Intelligence, University of the Basque Country (UPV/EHU) </li> <li> Donostia International Physics Center (DIPC) </li> </ul> |
| [Ignacio Arganda-Carreras](https://www.ikerbasque.net/es/ignacio-arganda-carreras) | Supervision, Implementation | <ul> <li> Dept. of Computer Science and Artificial Intelligence, University of the Basque Country (UPV/EHU) </li> <li> Donostia International Physics Center (DIPC) </li> <li> IKERBASQUE, Basque Foundation for Science </li> <li>Biofisika Institute </li> </ul> |
| [Arrate Muñoz-Barrutia](https://image.hggm.es/es/arrate-munoz) | Supervision | <ul> <li> Dept. de Bioingenieria, Universidad Carlos III de Madrid </li> </ul> |
## External collaborators
| Name | Role | Affiliations |
|----------------------------------------------------------------------------------------------------|-----------------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
| [Jesús Ángel Andrés-San Román](https://scholar.google.es/citations?user=OfDu4q4AAAAJ&hl=en&oi=sra) | Implementation | <ul> <li>Instituto de Biomedicina de Sevilla (IBiS), Hospital Universitario Virgen del Rocío/CSIC/Universidad de Sevilla and Dept. de Biología Celular, Facultad de Biología, Universidad de Sevilla </li> <li> Biomedical Network Research Centre on Neurodegenerative Diseases (CIBERNED) </li> </ul> |
| [Pedro Javier Gómez Gálvez](https://scholar.google.es/citations?user=aWeyQGUAAAAJ&hl=en&oi=sra) | Supervision, Implementation | <ul> <li>Instituto de Biomedicina de Sevilla (IBiS), Hospital Universitario Virgen del Rocío/CSIC/Universidad de Sevilla and Dept. de Biología Celular, Facultad de Biología, Universidad de Sevilla </li> <li> MRC Laboratory of Molecular Biology </li> <li> Department of Physiology, Development and Neuroscience, University of Cambridge </li> </ul> |
| [Luis M. Escudero](http://www.scutoids.es/) | Supervision | <ul> <li>Instituto de Biomedicina de Sevilla (IBiS), Hospital Universitario Virgen del Rocío/CSIC/Universidad de Sevilla and Dept. de Biología Celular, Facultad de Biología, Universidad de Sevilla </li> <li> Biomedical Network Research Centre on Neurodegenerative Diseases (CIBERNED) </li> </ul> |
| [Iván Hidalgo Cenalmor](https://henriqueslab.github.io/team/2023-03-16-IH/) | Implementation | <ul> <li> Optical cell biology group, Instituto Gulbenkian de Ciência, Oerias, Portugal </li> </lu> |
| [Donglai Wei](https://donglaiw.github.io/) | Supervision | <ul> <li>Boston College</li> </ul> |
| [Clément Caporal](https://scholar.google.com/citations?hl=en&user=R0fXJGUAAAAJ) | Implementation | <ul> <li>Laboratoire d’Optique et Biosciences, CNRS, Inserm, Ecole Polytechnique</li> </ul> |
| [Anatole Chessel](https://scholar.google.com/citations?hl=en&user=GC8aiVsAAAAJ) | Supervision | <ul> <li>Laboratoire d’Optique et Biosciences, CNRS, Inserm, Ecole Polytechnique</li> </ul> |
## Citation
Please note that BiaPy is based on a publication. If you use it successfully for your research please be so kind to cite our work:
```
Franco-Barranco, D., Andrés-San Román, J.A., Hidalgo-Cenalmor, I.,
Backová, L., González-Marfil, A., Caporal, C., Chessel, A., Gómez-Gálvez, P.,
Escudero, L.M., Wei, D., Muñoz-Barrutia, A. and Arganda-Carreras, I.
BiaPy: accessible deep learning on bioimages. Nat Methods 22, 1124–1126 (2025).
https://doi.org/10.1038/s41592-025-02699-y
```
Raw data
{
"_id": null,
"home_page": null,
"name": "biapy",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": "Daniel Franco-Barranco <daniel.franco@dipc.org>",
"keywords": "feed, reader, tutorial",
"author": null,
"author_email": "Daniel Franco-Barranco <daniel.franco@dipc.org>",
"download_url": "https://files.pythonhosted.org/packages/f8/71/a1388f39155b6552b5ee912e92bb460420a1978a53904cbfd6078cb9e489/biapy-3.6.4.tar.gz",
"platform": null,
"description": "\n\n# BiaPy: Accessible deep learning on bioimages\n\n<p align=\"left\">\n <a href=\"https://www.python.org/\">\n <img src=\"https://img.shields.io/badge/Python-3.10-yellow.svg\" /></a>\n <a href= \"https://pytorch.org/\">\n <img src=\"https://img.shields.io/badge/PyTorch-2.4-orange.svg\" /></a>\n <a href= \"https://pypi.org/project/biapy/\">\n <img src=\"https://img.shields.io/pypi/v/biapy.svg?color=green\" /></a>\n <a href= \"https://anaconda.org/conda-forge/biapy/\">\n <img src=\"https://anaconda.org/conda-forge/biapy/badges/version.svg\" /></a>\n <a href= \"https://github.com/BiaPyX/BiaPy/blob/master/LICENSE\">\n <img src=\"https://img.shields.io/badge/License-MIT-blue.svg\" /></a>\n <a href= \"https://biapy.readthedocs.io/en/latest/\">\n <img src=\"https://img.shields.io/badge/Doc-Latest-2BAF2B.svg\" /></a>\n <a href= \"https://www.nature.com/articles/s41592-025-02699-y\">\n <img src=\"https://img.shields.io/badge/Nature_Methods-Paper-bd2635.svg\" /></a>\n <a href= \"https://ieeexplore.ieee.org/abstract/document/10230593\">\n <img src=\"https://img.shields.io/badge/IEEE-Paper-00629B.svg\" /></a>\n</p>\n\n[](https://github.com/BiaPyX/BiaPy/actions/workflows/check_code_consistency.yml)\n[](https://github.com/BiaPyX/BiaPy/actions/workflows/upload_biapy_to_pypi.yml)\n[](https://github.com/BiaPyX/BiaPy/actions/workflows/create_release_container.yml)\n\n\ud83d\udd25**NEWS**\ud83d\udd25: BiaPy's paper is finally out in **Nature Methods**! Check it out at: [https://www.nature.com/articles/s41592-025-02699-y](https://www.nature.com/articles/s41592-025-02699-y)\n\nFor those who have no access to Nature Methods, here is the **preprint** at bioRxiv: [https://www.biorxiv.org/content/10.1101/2024.02.03.576026v3](https://www.biorxiv.org/content/10.1101/2024.02.03.576026v3)\n\n[BiaPy](https://biapyx.github.io) is an open source library and application that streamlines the use of common deep-learning workflows for a large variety of bioimage analysis tasks, including 2D and 3D [semantic segmentation](https://biapy.readthedocs.io/en/latest/workflows/semantic_segmentation.html), [instance segmentation](https://biapy.readthedocs.io/en/latest/workflows/instance_segmentation.html), [object detection](https://biapy.readthedocs.io/en/latest/workflows/detection.html), [image denoising](https://biapy.readthedocs.io/en/latest/workflows/denoising.html), [single image super-resolution](https://biapy.readthedocs.io/en/latest/workflows/super_resolution.html), [self-supervised learning](https://biapy.readthedocs.io/en/latest/workflows/self_supervision.html), [image classification](https://biapy.readthedocs.io/en/latest/workflows/classification.html) and [image to image translation](https://biapy.readthedocs.io/en/latest/workflows/image_to_image.html). \n \nBiaPy is a versatile platform designed to accommodate both proficient computer scientists and users less experienced in programming. It offers diverse and user-friendly access points to our workflows.\n\nThis repository is actively under development by the [BiaPy Team](https://biapyx.github.io/about/), a group of contributors from different research institutions.\n\n\n\n## Description videos\nFind a comprehensive overview of BiaPy and its functionality in the following videos:\n\n| [](https://www.youtube.com/watch?v=Gnm-VsZQ6Cc \"BiaPy: a ready-to-use library for Bioimage Analysis Pipelines\") <br> <span style=\"font-weight:normal\">BiaPy history and GUI demo at [RTmfm](https://rtmfm.cnrs.fr/en/) by Ignacio Arganda-Carreras and Daniel Franco-Barranco.</span> |\n|-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|\n| [](https://youtu.be/6eYtX-ySpc0 \"BiaPy: a ready-to-use library for Bioimage Analysis Pipelines\") <br> BiaPy presentation at [Virtual Pub of Euro-BioImaging](https://www.eurobioimaging.eu/) by Ignacio Arganda-Carreras. | | | | |\n\n## User interface\n\nYou can also use BiaPy through our graphical user interface (GUI).\n\n\n\n## Download BiaPy GUI for you OS\n\n- [Windows 64-bit](https://drive.google.com/uc?export=download&id=1iV0wzdFhpCpBCBgsameGyT3iFyQ6av5o) \n- [Linux 64-bit](https://drive.google.com/uc?export=download&id=13jllkLTR6S3yVZLRdMwhWUu7lq3HyJsD) \n- [macOS 64-bit](https://drive.google.com/uc?export=download&id=1fIpj9A8SWIN1fABEUAS--DNhOHzqSL7f) \n\nProject's page: [[BiaPy GUI](https://github.com/BiaPyX/BiaPy-GUI)]\n\n## Scientific publications using BiaPy\n\n| | |\n|--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|:-----------------------------------------------------------------------------------------------------------------------------------------------------------:|\n| ```L\u00f3pez-Cano, Daniel, et al. \"Characterizing Structure Formation through Instance Segmentation\" (2023).``` <br><br> This study presents a machine-learning framework to predict the formation of dark matter haloes from early universe density perturbations. Utilizing two neural networks, it distinguishes particles comprising haloes and groups them by membership. The framework accurately predicts halo masses and shapes, and compares favorably with N-body simulations. The open-source model could enhance analytical methods of structure formation by analyzing initial condition variations. ```BiaPy``` is used in the creation of the watershed approach. <br><br> [[Documentation (not yet)](https://biapy.readthedocs.io/en/latest)] [[Paper](https://arxiv.org/abs/2311.12110)] |  |\n| ```Franco-Barranco, Daniel, et al. \"Current Progress and Challenges in Large-scale 3D Mitochondria Instance Segmentation.\" (2023).``` <br><br> This paper reports the results of the MitoEM challenge on 3D instance segmentation of mitochondria in electron microscopy images, held in conjunction with IEEE-ISBI 2021. The paper discusses the top-performing methods, addresses ground truth errors, and proposes a new scoring system to improve segmentation evaluation. Despite progress, challenges remain in segmenting mitochondria with complex shapes, keeping the competition open for further submissions. ```BiaPy``` is used in the creation of the MitoEM challenge baseline (U2D-BC). <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/mitoem.html)] [[Paper](https://ieeexplore.ieee.org/document/10266382)] [[Toolbox](https://github.com/BiaPyX/TIMISE)] |  |\n| ```Backov\u00e1, Lenka, et al. \"Modeling Wound Healing Using Vector Quantized Variational Autoencoders and Transformers.\" 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI). IEEE, 2023.``` <br><br> This study focuses on time-lapse sequences of Drosophila embryos healing from laser-incised wounds. The researchers employ a two-stage approach involving a vector quantized variational autoencoder and an autoregressive transformer to model wound healing as a video prediction task. ```BiaPy``` is used in the creation of the wound segmentation masks. <br><br> [[Documentation](https://lenkaback.github.io/wound-healing-modeling/)] [[Paper](https://ieeexplore.ieee.org/document/10230571)] |  |\n| ```Andr\u00e9s-San Rom\u00e1n, Jes\u00fas A., et al. \"CartoCell, a high-content pipeline for 3D image analysis, unveils cell morphology patterns in epithelia.\" Cell Reports Methods (2023)``` <br><br> Combining deep learning and 3D imaging is crucial for high-content analysis. CartoCell is introduced, a method that accurately labels 3D epithelial cysts, enabling quantification of cellular features and mapping their distribution. It's adaptable to other epithelial tissues. CartoCell method is created using ```BiaPy```. <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/cartocell.html)] [[Paper](https://www.cell.com/cell-reports-methods/pdf/S2667-2375(23)00249-7.pdf)] | <table> <tr> <td> </td> <td> </td> </tr> </table> |\n| ```Franco-Barranco, Daniel, et al. \"Deep learning based domain adaptation for mitochondria segmentation on EM volumes.\" Computer Methods and Programs in Biomedicine 222 (2022): 106949.``` <br><br> This study addresses mitochondria segmentation across different datasets using three unsupervised domain adaptation approaches, including style transfer, self-supervised learning, and multi-task neural networks. To ensure robust generalization, a new training stopping criterion based on source domain morphological priors is proposed. ```BiaPy``` is used for the implementation of the Attention U-Net. <br><br> [[Documentation](https://github.com/danifranco/EM_domain_adaptation)] [[Paper](https://www.sciencedirect.com/science/article/pii/S0169260722003315)] |  |\n| ```Franco-Barranco, Daniel, et al. \"Stable deep neural network architectures for mitochondria segmentation on electron microscopy volumes.\" Neuroinformatics 20.2 (2022): 437-450.``` <br><br> Recent deep learning models have shown impressive performance in mitochondria segmentation, but often lack code and training details, affecting reproducibility. This study follows best practices, comprehensively comparing state-of-the-art architectures and variations of U-Net models for mitochondria segmentation, revealing their impact and stability. The research consistently achieves state-of-the-art results on various datasets, including EPFL Hippocampus, Lucchi++, and Kasthuri++. ```BiaPy``` is used for the implementation of the methods compared in the study. <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/stable.html)] [[Paper](https://link.springer.com/article/10.1007/s12021-021-09556-1)] |  |\n| ```Wei, Donglai, et al. \"Mitoem dataset: Large-scale 3d mitochondria instance segmentation from em images.\" International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer International Publishing, 2020.``` <br><br> Existing mitochondria segmentation datasets are small, raising questions about method robustness. The MitoEM dataset introduces larger 3D volumes with diverse mitochondria, challenging existing instance segmentation methods, highlighting the need for improved techniques. ```BiaPy``` is used in the creation of the MitoEM challenge baseline (U2D-BC). <br><br> [[Documentation](https://biapy.readthedocs.io/en/latest/tutorials/mitoem.html)] [[Paper](https://link.springer.com/chapter/10.1007/978-3-030-59722-1_7)] [[Challenge](https://mitoem.grand-challenge.org/)] |  |\n\n## Authors\n\n| Name | Role | Affiliations |\n|------------------------------------------------------------------------------------|------------------------------------------|---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|\n| [Daniel Franco-Barranco](https://BiaPyX.github.io/) | Creator, Implementation, Software design | <ul> <li> Dept. of Computer Science and Artificial Intelligence, University of the Basque Country (UPV/EHU) </li> <li> Donostia International Physics Center (DIPC) </li> </ul> |\n| [Lenka Backov\u00e1](https://www.biofisika.org/en/about/people/lenka-backova) | Implementation | <ul> <li> Biofisika Institute </li> </ul> |\n| [Aitor Gonzalez-Marfil](https://dipc.ehu.eus/es/dipc/personas/investigadores-en-formacion/aitor-gonzalez-marfil) | Implementation | <ul> <li> Dept. of Computer Science and Artificial Intelligence, University of the Basque Country (UPV/EHU) </li> <li> Donostia International Physics Center (DIPC) </li> </ul> |\n| [Ignacio Arganda-Carreras](https://www.ikerbasque.net/es/ignacio-arganda-carreras) | Supervision, Implementation | <ul> <li> Dept. of Computer Science and Artificial Intelligence, University of the Basque Country (UPV/EHU) </li> <li> Donostia International Physics Center (DIPC) </li> <li> IKERBASQUE, Basque Foundation for Science </li> <li>Biofisika Institute </li> </ul> |\n| [Arrate Mu\u00f1oz-Barrutia](https://image.hggm.es/es/arrate-munoz) | Supervision | <ul> <li> Dept. de Bioingenieria, Universidad Carlos III de Madrid </li> </ul> |\n\n\n## External collaborators\n\n| Name | Role | Affiliations |\n|----------------------------------------------------------------------------------------------------|-----------------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|\n| [Jes\u00fas \u00c1ngel Andr\u00e9s-San Rom\u00e1n](https://scholar.google.es/citations?user=OfDu4q4AAAAJ&hl=en&oi=sra) | Implementation | <ul> <li>Instituto de Biomedicina de Sevilla (IBiS), Hospital Universitario Virgen del Roc\u00edo/CSIC/Universidad de Sevilla and Dept. de Biolog\u00eda Celular, Facultad de Biolog\u00eda, Universidad de Sevilla </li> <li> Biomedical Network Research Centre on Neurodegenerative Diseases (CIBERNED) </li> </ul> |\n| [Pedro Javier G\u00f3mez G\u00e1lvez](https://scholar.google.es/citations?user=aWeyQGUAAAAJ&hl=en&oi=sra) | Supervision, Implementation | <ul> <li>Instituto de Biomedicina de Sevilla (IBiS), Hospital Universitario Virgen del Roc\u00edo/CSIC/Universidad de Sevilla and Dept. de Biolog\u00eda Celular, Facultad de Biolog\u00eda, Universidad de Sevilla </li> <li> MRC Laboratory of Molecular Biology </li> <li> Department of Physiology, Development and Neuroscience, University of Cambridge </li> </ul> |\n| [Luis M. Escudero](http://www.scutoids.es/) | Supervision | <ul> <li>Instituto de Biomedicina de Sevilla (IBiS), Hospital Universitario Virgen del Roc\u00edo/CSIC/Universidad de Sevilla and Dept. de Biolog\u00eda Celular, Facultad de Biolog\u00eda, Universidad de Sevilla </li> <li> Biomedical Network Research Centre on Neurodegenerative Diseases (CIBERNED) </li> </ul> |\n| [Iv\u00e1n Hidalgo Cenalmor](https://henriqueslab.github.io/team/2023-03-16-IH/) | Implementation | <ul> <li> Optical cell biology group, Instituto Gulbenkian de Ci\u00eancia, Oerias, Portugal </li> </lu> |\n| [Donglai Wei](https://donglaiw.github.io/) | Supervision | <ul> <li>Boston College</li> </ul> |\n| [Cl\u00e9ment Caporal](https://scholar.google.com/citations?hl=en&user=R0fXJGUAAAAJ) | Implementation | <ul> <li>Laboratoire d\u2019Optique et Biosciences, CNRS, Inserm, Ecole Polytechnique</li> </ul> |\n| [Anatole Chessel](https://scholar.google.com/citations?hl=en&user=GC8aiVsAAAAJ) | Supervision | <ul> <li>Laboratoire d\u2019Optique et Biosciences, CNRS, Inserm, Ecole Polytechnique</li> </ul> |\n\n## Citation\nPlease note that BiaPy is based on a publication. If you use it successfully for your research please be so kind to cite our work:\n```\nFranco-Barranco, D., Andr\u00e9s-San Rom\u00e1n, J.A., Hidalgo-Cenalmor, I.,\nBackov\u00e1, L., Gonz\u00e1lez-Marfil, A., Caporal, C., Chessel, A., G\u00f3mez-G\u00e1lvez, P.,\nEscudero, L.M., Wei, D., Mu\u00f1oz-Barrutia, A. and Arganda-Carreras, I.\nBiaPy: accessible deep learning on bioimages. Nat Methods 22, 1124\u20131126 (2025).\nhttps://doi.org/10.1038/s41592-025-02699-y\n``` \n",
"bugtrack_url": null,
"license": "MIT License\n \n Copyright (c) 2022 Daniel Franco-Barranco\n \n Permission is hereby granted, free of charge, to any person obtaining a copy\n of this software and associated documentation files (the \"Software\"), to deal\n in the Software without restriction, including without limitation the rights\n to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\n copies of the Software, and to permit persons to whom the Software is\n furnished to do so, subject to the following conditions:\n \n The above copyright notice and this permission notice shall be included in all\n copies or substantial portions of the Software.\n \n THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\n AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\n OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\n SOFTWARE.\n ",
"summary": "BiaPy: Bioimage analysis pipelines in Python",
"version": "3.6.4",
"project_urls": {
"Forum": "https://forum.image.sc/tag/biapy",
"Homepage": "https://biapyx.github.io",
"Source": "https://github.com/BiaPyX/BiaPy"
},
"split_keywords": [
"feed",
" reader",
" tutorial"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "a1060ca0eae0ddeae52bfa0a83299095f414de28d776d3f4f8b12eedc858f755",
"md5": "10435bab0b3dec12565b016b51213d85",
"sha256": "673c984d3d416ebb00d8be162319de668344b323f0eb1ff32db5c32e93f33ac6"
},
"downloads": -1,
"filename": "biapy-3.6.4-py3-none-any.whl",
"has_sig": false,
"md5_digest": "10435bab0b3dec12565b016b51213d85",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10",
"size": 531613,
"upload_time": "2025-08-15T07:17:25",
"upload_time_iso_8601": "2025-08-15T07:17:25.624301Z",
"url": "https://files.pythonhosted.org/packages/a1/06/0ca0eae0ddeae52bfa0a83299095f414de28d776d3f4f8b12eedc858f755/biapy-3.6.4-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "f871a1388f39155b6552b5ee912e92bb460420a1978a53904cbfd6078cb9e489",
"md5": "781c3d54f7917efa6f1a0bc0782e331d",
"sha256": "54de44a26b1423a04b6dbf55e0c2ecdc545c8b34394c787bc22cb876924a76fb"
},
"downloads": -1,
"filename": "biapy-3.6.4.tar.gz",
"has_sig": false,
"md5_digest": "781c3d54f7917efa6f1a0bc0782e331d",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 459259,
"upload_time": "2025-08-15T07:17:27",
"upload_time_iso_8601": "2025-08-15T07:17:27.317641Z",
"url": "https://files.pythonhosted.org/packages/f8/71/a1388f39155b6552b5ee912e92bb460420a1978a53904cbfd6078cb9e489/biapy-3.6.4.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-08-15 07:17:27",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "BiaPyX",
"github_project": "BiaPy",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "biapy"
}
