| Name | cehrbert-data JSON |
| Version |
0.1.1
 JSON
JSON |
| download |
| home_page | None |
| Summary | The Spark ETL tools for generating the CEHR-BERT and CEHR-GPT pre-training and finetuning data |
| upload_time | 2025-09-09 19:51:55 |
| maintainer | None |
| docs_url | None |
| author | None |
| requires_python | >=3.10.0 |
| license | MIT License |
| keywords |
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
# cehrbert_data
cehrbert_data is the ETL tool that generates the pretraining and finetuning datasets for CEHRbERT, which is a large language model developed for the structured EHR data, the work has been published
at https://proceedings.mlr.press/v158/pang21a.html.
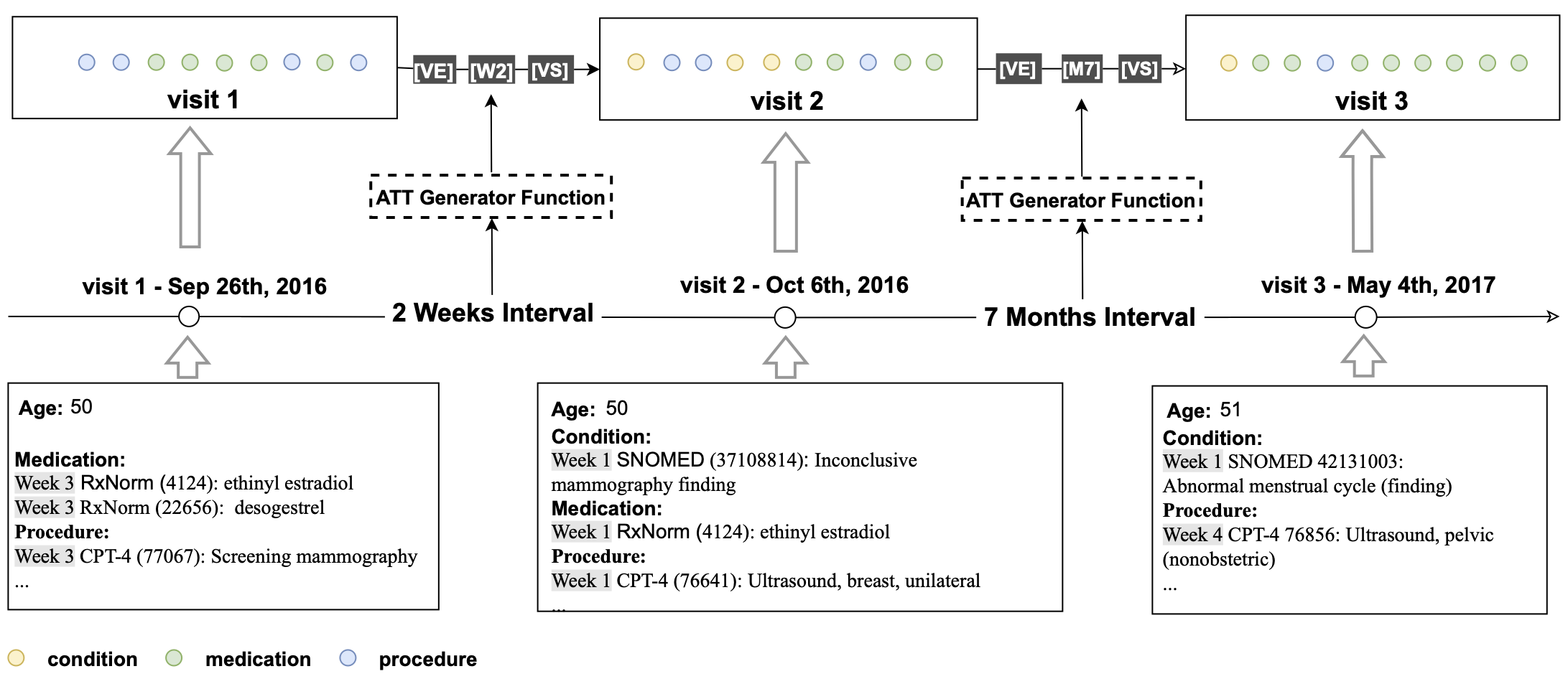
## Patient Representation
For each patient, all medical codes were aggregated and constructed into a sequence chronologically.
In order to incorporate temporal information, we inserted an artificial time token (ATT) between two neighboring visits
based on their time interval.
The following logic was used for creating ATTs based on the following time intervals between visits, if less than 28
days, ATTs take on the form of $W_n$ where n represents the week number ranging from 0-3 (e.g. $W_1$); 2) if between 28
days and 365 days, ATTs are in the form of **$M_n$** where n represents the month number ranging from 1-11 e.g $M_{11}$;
3) beyond 365 days then a **LT** (Long Term) token is inserted. In addition, we added two more special tokens — **VS**
and **VE** to represent the start and the end of a visit to explicitly define the visit segment, where all the
concepts
associated with the visit are subsumed by **VS** and **VE**.
## Pre-requisite
The project is built in python 3.10, and project dependency needs to be installed
Create a new Python virtual environment
```console
python3.10 -m venv .venv;
source .venv/bin/activate;
```
Build the project
```console
pip install -e .
```
Download [jtds-1.3.1.jar](jtds-1.3.1.jar) into the spark jars folder in the python environment
```console
cp jtds-1.3.1.jar .venv/lib/python3.10/site-packages/pyspark/jars/
```
## Instructions for Use
### 1. Download OMOP tables as parquet files
We created a spark app to download OMOP tables from SQL Server as parquet files. You need adjust the properties
in `db_properties.ini` to match with your database setup.
```console
PYTHONPATH=./: spark-submit tools/download_omop_tables.py -c db_properties.ini -tc person visit_occurrence condition_occurrence procedure_occurrence drug_exposure measurement observation_period concept concept_relationship concept_ancestor -o ~/Documents/omop_test/
```
We have prepared a synthea dataset with 1M patients for you to test, you could download it
at [omop_synthea.tar.gz](https://drive.google.com/file/d/1k7-cZACaDNw8A1JRI37mfMAhEErxKaQJ/view?usp=share_link)
```console
tar -xvf omop_synthea.tar ~/Document/omop_test/
```
### 2. Generate training data for CEHR-BERT
We order the patient events in chronological order and put all data points in a sequence. We insert artificial tokens
VS (visit start) and VE (visit end) to the start and the end of the visit. In addition, we insert artificial time
tokens (ATT) between visits to indicate the time interval between visits. This approach allows us to apply BERT to
structured EHR as-is.
The sequence can be seen conceptually as [VS] [V1] [VE] [ATT] [VS] [V2] [VE], where [V1] and [V2] represent a list of
concepts associated with those visits.
```console
PYTHONPATH=./: spark-submit spark_apps/generate_training_data.py -i ~/Documents/omop_test/ -o ~/Documents/omop_test/cehr-bert -tc condition_occurrence procedure_occurrence drug_exposure -d 1985-01-01 --is_new_patient_representation -iv
```
### 3. Generate hf readmission prediction task
If you don't have your own OMOP instance, we have provided a sample of patient sequence data generated using Synthea
at `sample/hf_readmissioon` in the repo
```console
PYTHONPATH=./:$PYTHONPATH spark-submit spark_apps/prediction_cohorts/hf_readmission.py -c hf_readmission -i ~/Documents/omop_test/ -o ~/Documents/omop_test/cehr-bert -dl 1985-01-01 -du 2020-12-31 -l 18 -u 100 -ow 360 -ps 0 -pw 30 --is_new_patient_representation
```
## Contact us
If you have any questions, feel free to contact us at CEHR-BERT@lists.cumc.columbia.edu
## Citation
Please acknowledge the following work in papers
Chao Pang, Xinzhuo Jiang, Krishna S. Kalluri, Matthew Spotnitz, RuiJun Chen, Adler
Perotte, and Karthik Natarajan. "Cehr-bert: Incorporating temporal information from
structured ehr data to improve prediction tasks." In Proceedings of Machine Learning for
Health, volume 158 of Proceedings of Machine Learning Research, pages 239–260. PMLR,
04 Dec 2021.
Raw data
{
"_id": null,
"home_page": null,
"name": "cehrbert-data",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10.0",
"maintainer_email": null,
"keywords": null,
"author": null,
"author_email": "Chao Pang <chaopang229@gmail.com>, Xinzhuo Jiang <xj2193@cumc.columbia.edu>, Krishna Kalluri <kk3326@cumc.columbia.edu>, Nishanth Parameshwar Pavinkurve <np2689@cumc.columbia.edu>, Karthik Natarajan <kn2174@cumc.columbia.edu>",
"download_url": "https://files.pythonhosted.org/packages/ba/3e/d202086313c171089a7faa1e20f816e76765c6aaa23946f627be51facf11/cehrbert_data-0.1.1.tar.gz",
"platform": null,
"description": "# cehrbert_data\n\ncehrbert_data is the ETL tool that generates the pretraining and finetuning datasets for CEHRbERT, which is a large language model developed for the structured EHR data, the work has been published\nat https://proceedings.mlr.press/v158/pang21a.html.\n\n## Patient Representation\nFor each patient, all medical codes were aggregated and constructed into a sequence chronologically.\nIn order to incorporate temporal information, we inserted an artificial time token (ATT) between two neighboring visits\nbased on their time interval.\nThe following logic was used for creating ATTs based on the following time intervals between visits, if less than 28\ndays, ATTs take on the form of $W_n$ where n represents the week number ranging from 0-3 (e.g. $W_1$); 2) if between 28\ndays and 365 days, ATTs are in the form of **$M_n$** where n represents the month number ranging from 1-11 e.g $M_{11}$;\n\n3) beyond 365 days then a **LT** (Long Term) token is inserted. In addition, we added two more special tokens \u2014 **VS**\n and **VE** to represent the start and the end of a visit to explicitly define the visit segment, where all the\n concepts\n associated with the visit are subsumed by **VS** and **VE**.\n\n\n\n## Pre-requisite\nThe project is built in python 3.10, and project dependency needs to be installed\n\nCreate a new Python virtual environment\n\n```console\npython3.10 -m venv .venv;\nsource .venv/bin/activate;\n```\n\nBuild the project\n\n```console\npip install -e .\n```\n\nDownload [jtds-1.3.1.jar](jtds-1.3.1.jar) into the spark jars folder in the python environment\n```console\ncp jtds-1.3.1.jar .venv/lib/python3.10/site-packages/pyspark/jars/\n```\n## Instructions for Use\n\n### 1. Download OMOP tables as parquet files\n\nWe created a spark app to download OMOP tables from SQL Server as parquet files. You need adjust the properties\nin `db_properties.ini` to match with your database setup.\n\n```console\nPYTHONPATH=./: spark-submit tools/download_omop_tables.py -c db_properties.ini -tc person visit_occurrence condition_occurrence procedure_occurrence drug_exposure measurement observation_period concept concept_relationship concept_ancestor -o ~/Documents/omop_test/\n```\n\nWe have prepared a synthea dataset with 1M patients for you to test, you could download it\nat [omop_synthea.tar.gz](https://drive.google.com/file/d/1k7-cZACaDNw8A1JRI37mfMAhEErxKaQJ/view?usp=share_link)\n\n```console\ntar -xvf omop_synthea.tar ~/Document/omop_test/\n```\n\n### 2. Generate training data for CEHR-BERT\nWe order the patient events in chronological order and put all data points in a sequence. We insert artificial tokens\nVS (visit start) and VE (visit end) to the start and the end of the visit. In addition, we insert artificial time\ntokens (ATT) between visits to indicate the time interval between visits. This approach allows us to apply BERT to\nstructured EHR as-is.\nThe sequence can be seen conceptually as [VS] [V1] [VE] [ATT] [VS] [V2] [VE], where [V1] and [V2] represent a list of\nconcepts associated with those visits.\n\n```console\nPYTHONPATH=./: spark-submit spark_apps/generate_training_data.py -i ~/Documents/omop_test/ -o ~/Documents/omop_test/cehr-bert -tc condition_occurrence procedure_occurrence drug_exposure -d 1985-01-01 --is_new_patient_representation -iv\n```\n### 3. Generate hf readmission prediction task\nIf you don't have your own OMOP instance, we have provided a sample of patient sequence data generated using Synthea\nat `sample/hf_readmissioon` in the repo\n\n```console\nPYTHONPATH=./:$PYTHONPATH spark-submit spark_apps/prediction_cohorts/hf_readmission.py -c hf_readmission -i ~/Documents/omop_test/ -o ~/Documents/omop_test/cehr-bert -dl 1985-01-01 -du 2020-12-31 -l 18 -u 100 -ow 360 -ps 0 -pw 30 --is_new_patient_representation\n```\n\n## Contact us\nIf you have any questions, feel free to contact us at CEHR-BERT@lists.cumc.columbia.edu\n\n## Citation\nPlease acknowledge the following work in papers\n\nChao Pang, Xinzhuo Jiang, Krishna S. Kalluri, Matthew Spotnitz, RuiJun Chen, Adler\nPerotte, and Karthik Natarajan. \"Cehr-bert: Incorporating temporal information from\nstructured ehr data to improve prediction tasks.\" In Proceedings of Machine Learning for\nHealth, volume 158 of Proceedings of Machine Learning Research, pages 239\u2013260. PMLR,\n04 Dec 2021.\n",
"bugtrack_url": null,
"license": "MIT License",
"summary": "The Spark ETL tools for generating the CEHR-BERT and CEHR-GPT pre-training and finetuning data",
"version": "0.1.1",
"project_urls": {
"Homepage": "https://github.com/knatarajan-lab/cehrbert_data"
},
"split_keywords": [],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "a709acf9400acb846c5a092dda9475e213073945e65a177c14337bd008793ff8",
"md5": "bb14a20166b533c9b337ea0de2901ef2",
"sha256": "929d2efbad74f894666b07b177ad9adbab9285a9d44deee262eb81c334e64937"
},
"downloads": -1,
"filename": "cehrbert_data-0.1.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "bb14a20166b533c9b337ea0de2901ef2",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10.0",
"size": 95213,
"upload_time": "2025-09-09T19:51:54",
"upload_time_iso_8601": "2025-09-09T19:51:54.451500Z",
"url": "https://files.pythonhosted.org/packages/a7/09/acf9400acb846c5a092dda9475e213073945e65a177c14337bd008793ff8/cehrbert_data-0.1.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "ba3ed202086313c171089a7faa1e20f816e76765c6aaa23946f627be51facf11",
"md5": "32006970a478b8b3cc17040e1406a7c2",
"sha256": "5df2ab5f3d625a532a1e386fa04197b5ec6a8845ae6f8788ea9405bd34988c2f"
},
"downloads": -1,
"filename": "cehrbert_data-0.1.1.tar.gz",
"has_sig": false,
"md5_digest": "32006970a478b8b3cc17040e1406a7c2",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10.0",
"size": 425202,
"upload_time": "2025-09-09T19:51:55",
"upload_time_iso_8601": "2025-09-09T19:51:55.853797Z",
"url": "https://files.pythonhosted.org/packages/ba/3e/d202086313c171089a7faa1e20f816e76765c6aaa23946f627be51facf11/cehrbert_data-0.1.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-09-09 19:51:55",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "knatarajan-lab",
"github_project": "cehrbert_data",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "cehrbert-data"
}
