[](https://cfutils.readthedocs.io/en/latest/?badge=latest)
[](https://travis-ci.org/y9c/cfutils)
[](https://pypi.python.org/pypi/cfutils)
[](https://pepy.tech/project/cfutils)
**Chromatogram File Utils**
For Sanger sequencing data visualizing, alignment, mutation calling, and trimming etc.
## Demo
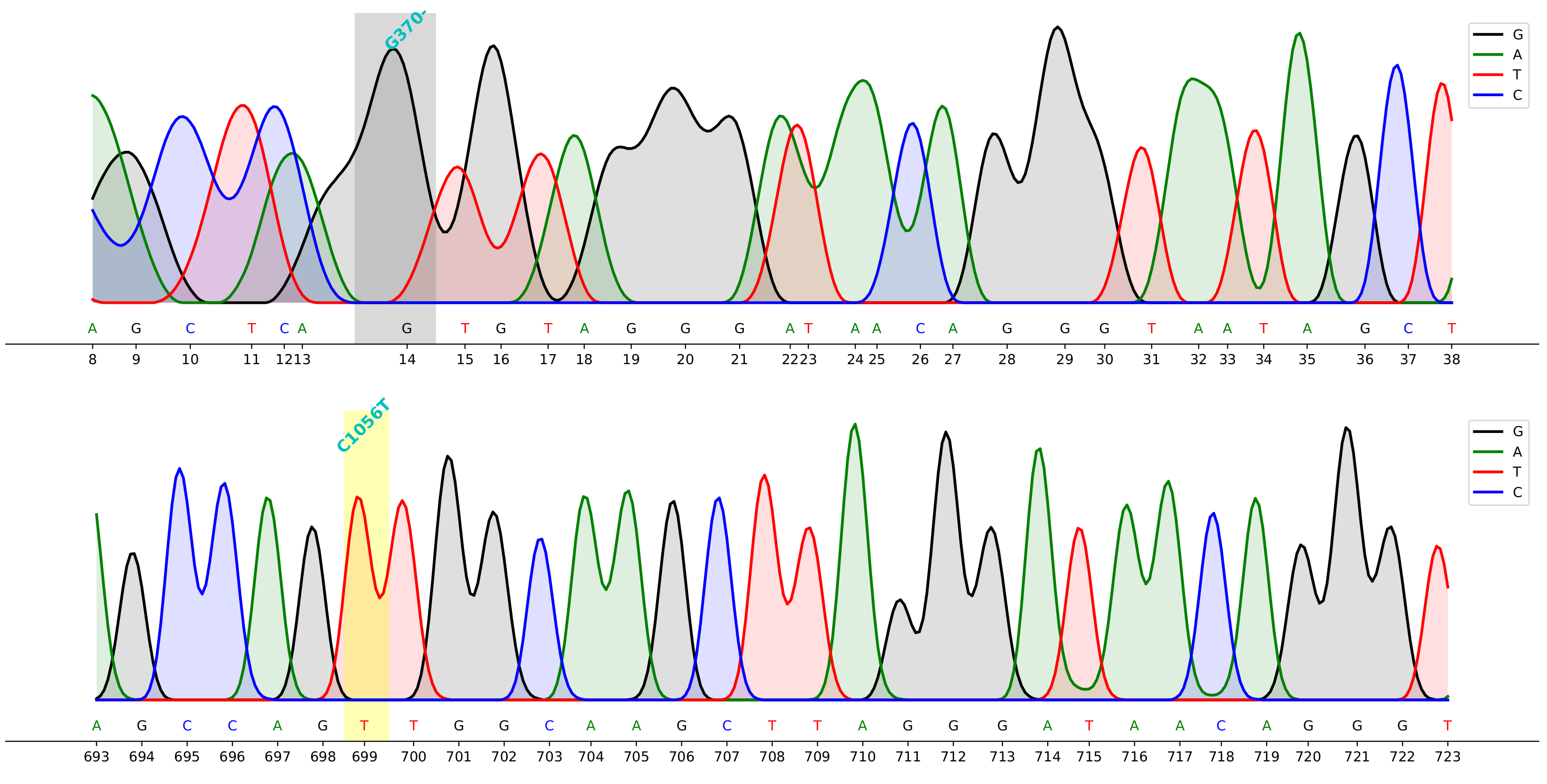
> command to generate the demo above
```bash
cfutils mut --query ./data/B5-M13R_B07.ab1 --subject ./data/ref.fa --outdir ./data/ --plot
```
## How to use?
- You can have mutation detection and visualization in one step using the command line.
```bash
cfutils mut --help
```
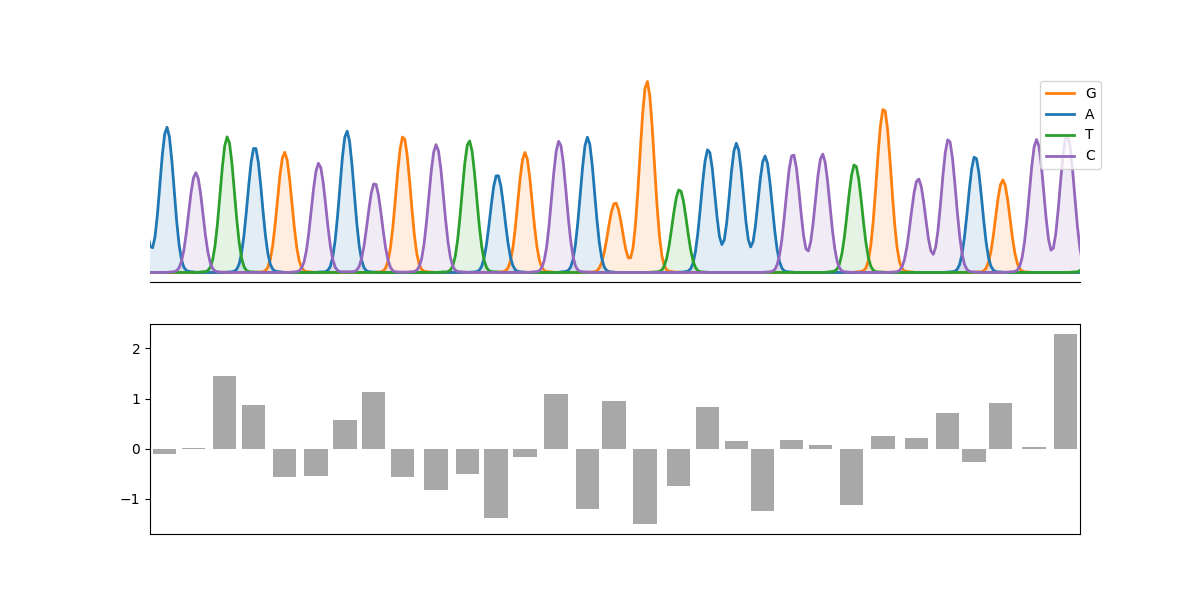
- You can also integrate the result matplotlib figures and use it as a python module.
An example:
```python
import matplotlib.pyplot as plt
import numpy as np
from cfutils.parser import parse_abi
from cfutils.show import plot_chromatograph
seq = parse_abi("./data/B5-M13R_B07.ab1")
peaks = seq.annotations["peak positions"][100:131]
fig, axes = plt.subplots(2, 1, figsize=(12, 6), sharex=True)
plot_chromatograph(
seq,
region=(100, 130),
ax=axes[0],
show_bases=True,
show_positions=True,
color_map=dict(zip("ATGC", ["C0", "C2", "C1", "C4"])),
)
axes[1].bar(peaks, np.random.randn(len(peaks)), color="0.66")
plt.show()
```
## How to install?
### form pypi
_(use this way ONLY, if you don't know what's going on)_
```bash
pip install --user cfutils
```
### manipulate the source code
- clone from github
```bash
git clone git@github.com:y9c/cfutils.git
```
- install the dependence
```bash
make init
```
- do unittest
```bash
make test
```
## ChangeLog
- Reverse completement the chromatogram file. (Inspired by Snapgene)
- build as python package for pypi
- fix bug that highlighting wrong base
- replace blastn with buildin python aligner
## TODO
- [ ] call mutation by alignment and plot Chromatogram graphic
- [ ] add a doc
- [x] change xaxis by peak location
- [ ] fix bug that chromatogram switch pos after trim
- [x] wrap as a cli app
- [ ] return quality score in output
- [ ] fix issue that selected base is not in the middle
- [ ] fix plot_chromatograph rendering bug
- [ ] add projection feature to make align and assemble possible
Raw data
{
"_id": null,
"home_page": "https://github.com/yech1990/cfutils",
"name": "cfutils",
"maintainer": null,
"docs_url": null,
"requires_python": "<4.0,>=3.8",
"maintainer_email": null,
"keywords": "DNA, mutation, chromatogram, biology",
"author": "Ye Chang",
"author_email": "yech1990@gmail.com",
"download_url": "https://files.pythonhosted.org/packages/07/b5/32a5a55014c9575cfcc73b25a6e0a5e570b12ad4716df741f73f9654e778/cfutils-0.0.0.dev60.tar.gz",
"platform": null,
"description": "[](https://cfutils.readthedocs.io/en/latest/?badge=latest)\n[](https://travis-ci.org/y9c/cfutils)\n[](https://pypi.python.org/pypi/cfutils)\n[](https://pepy.tech/project/cfutils)\n\n**Chromatogram File Utils**\n\nFor Sanger sequencing data visualizing, alignment, mutation calling, and trimming etc.\n\n## Demo\n\n\n\n> command to generate the demo above\n\n```bash\ncfutils mut --query ./data/B5-M13R_B07.ab1 --subject ./data/ref.fa --outdir ./data/ --plot\n```\n\n## How to use?\n\n- You can have mutation detection and visualization in one step using the command line.\n\n```bash\ncfutils mut --help\n```\n\n- You can also integrate the result matplotlib figures and use it as a python module.\n\nAn example:\n\n```python\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom cfutils.parser import parse_abi\nfrom cfutils.show import plot_chromatograph\n\nseq = parse_abi(\"./data/B5-M13R_B07.ab1\")\npeaks = seq.annotations[\"peak positions\"][100:131]\n\nfig, axes = plt.subplots(2, 1, figsize=(12, 6), sharex=True)\nplot_chromatograph(\n seq,\n region=(100, 130),\n ax=axes[0],\n show_bases=True,\n show_positions=True,\n color_map=dict(zip(\"ATGC\", [\"C0\", \"C2\", \"C1\", \"C4\"])),\n)\naxes[1].bar(peaks, np.random.randn(len(peaks)), color=\"0.66\")\nplt.show()\n```\n\n\n\n## How to install?\n\n### form pypi\n\n_(use this way ONLY, if you don't know what's going on)_\n\n```bash\npip install --user cfutils\n```\n\n### manipulate the source code\n\n- clone from github\n\n```bash\ngit clone git@github.com:y9c/cfutils.git\n```\n\n- install the dependence\n\n```bash\nmake init\n```\n\n- do unittest\n\n```bash\nmake test\n```\n\n## ChangeLog\n\n- Reverse completement the chromatogram file. (Inspired by Snapgene)\n- build as python package for pypi\n- fix bug that highlighting wrong base\n- replace blastn with buildin python aligner\n\n## TODO\n\n- [ ] call mutation by alignment and plot Chromatogram graphic\n- [ ] add a doc\n- [x] change xaxis by peak location\n- [ ] fix bug that chromatogram switch pos after trim\n- [x] wrap as a cli app\n- [ ] return quality score in output\n- [ ] fix issue that selected base is not in the middle\n- [ ] fix plot_chromatograph rendering bug\n\n- [ ] add projection feature to make align and assemble possible\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Chromatogram File Utils",
"version": "0.0.0.dev60",
"project_urls": {
"Documentation": "https://cf.readthedocs.io/",
"Homepage": "https://github.com/yech1990/cfutils",
"Repository": "https://github.com/yech1990/cfutils"
},
"split_keywords": [
"dna",
" mutation",
" chromatogram",
" biology"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "ef9917b7d707964411fd1789929edb7171bb3b886e8f4de3556ecbb79e96852f",
"md5": "275b4f172477cf720e70ca943db1a1ca",
"sha256": "63a3e080dd98fead8d12a343256acc3db405eeaf411e21f6ad72a0803f72c531"
},
"downloads": -1,
"filename": "cfutils-0.0.0.dev60-py3-none-any.whl",
"has_sig": false,
"md5_digest": "275b4f172477cf720e70ca943db1a1ca",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": "<4.0,>=3.8",
"size": 15430,
"upload_time": "2024-03-29T08:40:07",
"upload_time_iso_8601": "2024-03-29T08:40:07.562321Z",
"url": "https://files.pythonhosted.org/packages/ef/99/17b7d707964411fd1789929edb7171bb3b886e8f4de3556ecbb79e96852f/cfutils-0.0.0.dev60-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "07b532a5a55014c9575cfcc73b25a6e0a5e570b12ad4716df741f73f9654e778",
"md5": "7377bf76b09ecd6ba127f63a4bb160f8",
"sha256": "2d3e6dcd054358aa0cf8f03e015b9e5fae2c5b63fc009136d01483eb4a7e6e2a"
},
"downloads": -1,
"filename": "cfutils-0.0.0.dev60.tar.gz",
"has_sig": false,
"md5_digest": "7377bf76b09ecd6ba127f63a4bb160f8",
"packagetype": "sdist",
"python_version": "source",
"requires_python": "<4.0,>=3.8",
"size": 13413,
"upload_time": "2024-03-29T08:40:08",
"upload_time_iso_8601": "2024-03-29T08:40:08.946192Z",
"url": "https://files.pythonhosted.org/packages/07/b5/32a5a55014c9575cfcc73b25a6e0a5e570b12ad4716df741f73f9654e778/cfutils-0.0.0.dev60.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-03-29 08:40:08",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "yech1990",
"github_project": "cfutils",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "cfutils"
}
