| Name | crisprzip JSON |
| Version |
1.2.1
 JSON
JSON |
| download |
| home_page | None |
| Summary | A kinetic model of CRISPR-Cas target recognition. |
| upload_time | 2025-07-31 13:37:25 |
| maintainer | None |
| docs_url | None |
| author | None |
| requires_python | <3.13,>=3.10 |
| license | MIT License
Copyright (c) 2024 Hidde Offerhaus
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE. |
| keywords |
crispr-cas9
crispr
bioinformatics
genome editing
genetic engineering
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
# CRISPRzip
[](https://opensource.org/licenses/MIT)


Welcome to the codebase of CRISPRzip from the [Depken Lab](https://depkenlab.tudelft.nl/) at TU
Delft.
## About the project
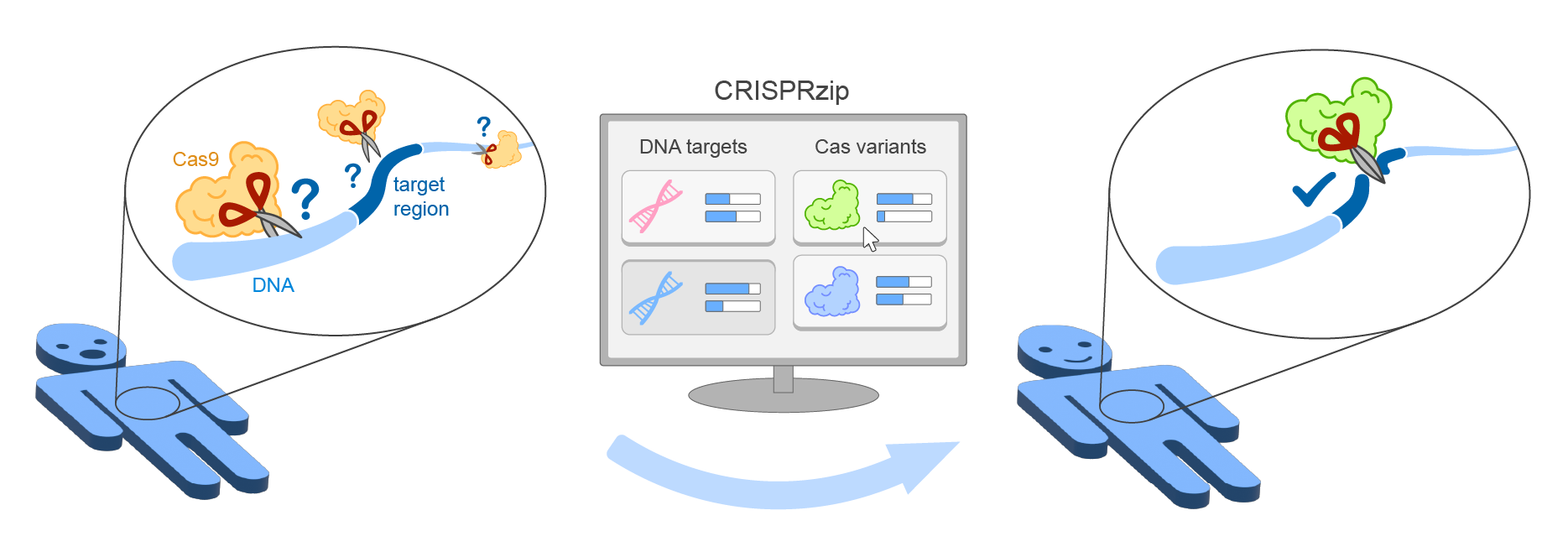
CRISPRzip is a physics-based model to study the target
recognition dynamics of CRISPR-associated nucleases like Cas9
([Eslami-Mossalam, 2022](#references)). Their interactions with target DNA is represented
as an energy landscape, with which you can simulate binding and cleavage
kinetics. The parameters have been obtained by machine learning on
high-throughput data. CRISPRzip makes quantitative predictions of on-target
efficiency and off-target risks of different guide RNAs.
With CRISPRzip, we hope to contribute to assessing
the risks that come with particular choices in CRISPR application, and as such
contribute to the development of safe gene editing technology.
### References
Eslami-Mossallam B et al. (2022) *A kinetic model predicts SpCas9 activity,
improves off-target classification, and reveals the physical basis of
targeting fidelity.* Nature Communications.
[10.1038/s41467-022-28994-2](https://doi.org/10.1038/s41467-022-28994-2)
## Installation
CRISPRzip is available on [PyPi](https://pypi.org/) and can be installed
with [pip](https://pip.pypa.io/en/stable/).
### User installation
Although CRISPRzip can be directly installed from pip, creating a virtual
environment makes it easier to manage dependencies. Below you can see some
intructions on how to generate a virtual environment with
[venv](https://docs.python.org/3/library/venv.html) assuming you already
have python installed in a bash-like terminal.
```shell
python -m venv crisprzip-venv
source crisprzip-venv/bin/activate
pip install crisprzip
```
### Developer installation
To be able to make changes and contributions to CRISPRzip, you will need to
get your own copy of the source code and install software dependencies on
your own. Assuming you have a python and git installed in a bash-like
terminal, the installation process can be done with the following
instructions.
```shell
git clone https://github.com/hiddeoff/crisprzip.git
cd crisprzip
python -m venv crisprzip-venv
source crisprzip-venv/bin/activate
pip install -e .
```
Please use our
[contributing guidelines](https://github.com/hiddeoff/crisprzip/blob/main/CONTRIBUTING.md)
if you would like us to consider your developments in a future CRISPRzip
release
### Verifying installation
CRISPRzip development includes a cross-platform compatibility and test
[workflow](.github/workflows/compatibility-test.yml). If you would like to
verify your local installation, please follow the Developer Installation
instructions, then run the following test.
```shell
source crisprzip-venv/bin/activate
pip install -e '.[tests]' # installs pytest and pandas
pytest tests/cleavage_binding_prediction/test_cleavage_binding_prediction.py -v
```
You can also follow the user installation and execute the code in the
Usage section below.
## Usage
CRISPRzip makes predictions about cleavage and binding activity on on- and
off-targets. First, you define the protospacer and target sequence, and then,
you can predict the fraction cleaved or bound.
```python
# 1. load parameter set
from crisprzip.kinetics import load_landscape
searcher = load_landscape("sequence_params")
# 2. define Cas9, gRNA and DNA target
searchertargetcomplex = searcher.probe_sequence(
protospacer = "AGACGCATAAAGATGAGACGCTGG",
target_seq = "AGACCCATTAAGATGAGACGCGGG", # A13T G17C
)
# 3. predict activity
f_clv = searchertargetcomplex.get_cleaved_fraction(
time=600, # 10 minutes
on_rate=1E-1
)
f_bnd = searchertargetcomplex.get_bound_fraction(
time=600, # 10 minutes
on_rate=1E-1
)
# 4. format output
print(f"After 10 minutes, the target (A13T G17C) is ...")
print(f"- cleaved for {100 * f_clv:.1f}% by Cas9")
print(f" or ")
print(f"- bound for {100 * f_bnd:.1f}% by dCas9")
```
Output:
```
After 10 minutes, the target (A13T G17C) is ...
- cleaved for 10.5% by Cas9
or
- bound for 94.2% by dCas9
```
See the [tutorial](examples/tutorial.ipynb) or the
[docs](https://hiddeoff.github.io/crisprzip/) for more examples how to explore
sequence, time and concentration dependency.
## Contributing
We encourage contributions in any form - reporting bugs, suggesting features,
drafting code changes. Read our [Contributing guidelines](./CONTRIBUTING.md) and
our [Code of Conduct](./CODE_OF_CONDUCT.md).
## Acknowledgements
Many thanks to [Elviss Dvinskis](https://github.com/edvinskis),
[Raúl Ortiz](https://github.com/rortizmerino) and [Aysun Urhan](https://github.com/aysunrhn)
from the [DCC team at TU Delft](https://www.tudelft.nl/en/library/support/library-for-researchers/setting-up-research/dcc)
for their support to get this package released!
## Waiver
Technische Universiteit Delft hereby disclaims all copyright interest in the
program “CRISPRzip” (a physics-based CRISPR activity predictor)
written by the Author(s).
Paulien Herder, Dean of Applied Sciences
(c) 2024, Hidde Offerhaus, Delft, The Netherlands.
Raw data
{
"_id": null,
"home_page": null,
"name": "crisprzip",
"maintainer": null,
"docs_url": null,
"requires_python": "<3.13,>=3.10",
"maintainer_email": "HS Offerhaus <hsofferhaus@gmail.com>",
"keywords": "crispr-cas9, crispr, bioinformatics, genome editing, genetic engineering",
"author": null,
"author_email": "HS Offerhaus <hsofferhaus@gmail.com>",
"download_url": "https://files.pythonhosted.org/packages/67/93/5358b855b35176aacea13e166e969307f1f27f363d30f243763cd89f5773/crisprzip-1.2.1.tar.gz",
"platform": null,
"description": "# CRISPRzip\r\n[](https://opensource.org/licenses/MIT)\r\n\r\n \r\n\r\nWelcome to the codebase of CRISPRzip from the [Depken Lab](https://depkenlab.tudelft.nl/) at TU\r\nDelft.\r\n\r\n## About the project\r\n\r\n\r\nCRISPRzip is a physics-based model to study the target \r\nrecognition dynamics of CRISPR-associated nucleases like Cas9\r\n([Eslami-Mossalam, 2022](#references)). Their interactions with target DNA is represented \r\nas an energy landscape, with which you can simulate binding and cleavage\r\nkinetics. The parameters have been obtained by machine learning on \r\nhigh-throughput data. CRISPRzip makes quantitative predictions of on-target \r\nefficiency and off-target risks of different guide RNAs.\r\n\r\nWith CRISPRzip, we hope to contribute to assessing\r\nthe risks that come with particular choices in CRISPR application, and as such\r\ncontribute to the development of safe gene editing technology.\r\n\r\n### References\r\nEslami-Mossallam B et al. (2022) *A kinetic model predicts SpCas9 activity,\r\nimproves off-target classification, and reveals the physical basis of\r\ntargeting fidelity.* Nature Communications.\r\n[10.1038/s41467-022-28994-2](https://doi.org/10.1038/s41467-022-28994-2)\r\n\r\n## Installation\r\nCRISPRzip is available on [PyPi](https://pypi.org/) and can be installed \r\nwith [pip](https://pip.pypa.io/en/stable/).\r\n\r\n### User installation\r\nAlthough CRISPRzip can be directly installed from pip, creating a virtual \r\nenvironment makes it easier to manage dependencies. Below you can see some \r\nintructions on how to generate a virtual environment with \r\n[venv](https://docs.python.org/3/library/venv.html) assuming you already \r\nhave python installed in a bash-like terminal.\r\n\r\n```shell\r\npython -m venv crisprzip-venv\r\nsource crisprzip-venv/bin/activate\r\npip install crisprzip\r\n```\r\n\r\n### Developer installation\r\nTo be able to make changes and contributions to CRISPRzip, you will need to \r\nget your own copy of the source code and install software dependencies on \r\nyour own. Assuming you have a python and git installed in a bash-like \r\nterminal, the installation process can be done with the following \r\ninstructions.\r\n\r\n```shell\r\ngit clone https://github.com/hiddeoff/crisprzip.git\r\ncd crisprzip\r\npython -m venv crisprzip-venv\r\nsource crisprzip-venv/bin/activate\r\npip install -e .\r\n```\r\n\r\nPlease use our \r\n[contributing guidelines](https://github.com/hiddeoff/crisprzip/blob/main/CONTRIBUTING.md)\r\n if you would like us to consider your developments in a future CRISPRzip \r\nrelease\r\n\r\n### Verifying installation\r\nCRISPRzip development includes a cross-platform compatibility and test \r\n[workflow](.github/workflows/compatibility-test.yml). If you would like to \r\nverify your local installation, please follow the Developer Installation \r\ninstructions, then run the following test.\r\n\r\n```shell\r\nsource crisprzip-venv/bin/activate\r\npip install -e '.[tests]' # installs pytest and pandas\r\npytest tests/cleavage_binding_prediction/test_cleavage_binding_prediction.py -v\r\n```\r\n\r\nYou can also follow the user installation and execute the code in the \r\nUsage section below.\r\n\r\n## Usage\r\nCRISPRzip makes predictions about cleavage and binding activity on on- and\r\noff-targets. First, you define the protospacer and target sequence, and then,\r\nyou can predict the fraction cleaved or bound.\r\n\r\n```python\r\n# 1. load parameter set\r\nfrom crisprzip.kinetics import load_landscape\r\nsearcher = load_landscape(\"sequence_params\")\r\n\r\n# 2. define Cas9, gRNA and DNA target\r\nsearchertargetcomplex = searcher.probe_sequence(\r\n protospacer = \"AGACGCATAAAGATGAGACGCTGG\",\r\n target_seq = \"AGACCCATTAAGATGAGACGCGGG\", # A13T G17C\r\n)\r\n\r\n# 3. predict activity\r\nf_clv = searchertargetcomplex.get_cleaved_fraction(\r\n time=600, # 10 minutes\r\n on_rate=1E-1\r\n)\r\nf_bnd = searchertargetcomplex.get_bound_fraction(\r\n time=600, # 10 minutes\r\n on_rate=1E-1\r\n)\r\n\r\n# 4. format output\r\nprint(f\"After 10 minutes, the target (A13T G17C) is ...\")\r\nprint(f\"- cleaved for {100 * f_clv:.1f}% by Cas9\")\r\nprint(f\" or \")\r\nprint(f\"- bound for {100 * f_bnd:.1f}% by dCas9\")\r\n```\r\nOutput:\r\n```\r\nAfter 10 minutes, the target (A13T G17C) is ...\r\n- cleaved for 10.5% by Cas9\r\n or \r\n- bound for 94.2% by dCas9\r\n```\r\n\r\nSee the [tutorial](examples/tutorial.ipynb) or the\r\n[docs](https://hiddeoff.github.io/crisprzip/) for more examples how to explore \r\nsequence, time and concentration dependency.\r\n\r\n## Contributing\r\nWe encourage contributions in any form - reporting bugs, suggesting features,\r\ndrafting code changes. Read our [Contributing guidelines](./CONTRIBUTING.md) and \r\nour [Code of Conduct](./CODE_OF_CONDUCT.md).\r\n\r\n## Acknowledgements\r\nMany thanks to [Elviss Dvinskis](https://github.com/edvinskis),\r\n[Ra\u00fal Ortiz](https://github.com/rortizmerino) and [Aysun Urhan](https://github.com/aysunrhn)\r\nfrom the [DCC team at TU Delft](https://www.tudelft.nl/en/library/support/library-for-researchers/setting-up-research/dcc)\r\nfor their support to get this package released!\r\n\r\n## Waiver\r\nTechnische Universiteit Delft hereby disclaims all copyright interest in the\r\nprogram \u201cCRISPRzip\u201d (a physics-based CRISPR activity predictor)\r\nwritten by the Author(s).\r\nPaulien Herder, Dean of Applied Sciences\r\n\r\n(c) 2024, Hidde Offerhaus, Delft, The Netherlands.\r\n",
"bugtrack_url": null,
"license": "MIT License\r\n \r\n Copyright (c) 2024 Hidde Offerhaus\r\n \r\n Permission is hereby granted, free of charge, to any person obtaining a copy\r\n of this software and associated documentation files (the \"Software\"), to deal\r\n in the Software without restriction, including without limitation the rights\r\n to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\r\n copies of the Software, and to permit persons to whom the Software is\r\n furnished to do so, subject to the following conditions:\r\n \r\n The above copyright notice and this permission notice shall be included in all\r\n copies or substantial portions of the Software.\r\n \r\n THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\r\n IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\r\n FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\r\n AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\r\n LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\r\n OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\r\n SOFTWARE.",
"summary": "A kinetic model of CRISPR-Cas target recognition.",
"version": "1.2.1",
"project_urls": {
"Documentation": "https://hiddeoff.github.io/crisprzip/",
"Homepage": "https://github.com/hiddeoff/crisprzip",
"Source": "https://github.com/hiddeoff/crisprzip"
},
"split_keywords": [
"crispr-cas9",
" crispr",
" bioinformatics",
" genome editing",
" genetic engineering"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "a1628f18461ecfa825077d1635e8ac2bd9581bec783cf35335124a3e690f2259",
"md5": "de5e87c64cd4c6662c80a18737ba05c8",
"sha256": "96fea5bde2abbb1a7a8bc797122cc94c7aff3b1a3ef52edf7b796dd5470be50d"
},
"downloads": -1,
"filename": "crisprzip-1.2.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "de5e87c64cd4c6662c80a18737ba05c8",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": "<3.13,>=3.10",
"size": 48222,
"upload_time": "2025-07-31T13:37:24",
"upload_time_iso_8601": "2025-07-31T13:37:24.049423Z",
"url": "https://files.pythonhosted.org/packages/a1/62/8f18461ecfa825077d1635e8ac2bd9581bec783cf35335124a3e690f2259/crisprzip-1.2.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "67935358b855b35176aacea13e166e969307f1f27f363d30f243763cd89f5773",
"md5": "5eb9184040928064b6ec5b5d3a6b0891",
"sha256": "ee222f3539ef2af7d64b009d9047d014a04a00ed6f8b9c20d1f5406dbc9b5ad9"
},
"downloads": -1,
"filename": "crisprzip-1.2.1.tar.gz",
"has_sig": false,
"md5_digest": "5eb9184040928064b6ec5b5d3a6b0891",
"packagetype": "sdist",
"python_version": "source",
"requires_python": "<3.13,>=3.10",
"size": 48654,
"upload_time": "2025-07-31T13:37:25",
"upload_time_iso_8601": "2025-07-31T13:37:25.022869Z",
"url": "https://files.pythonhosted.org/packages/67/93/5358b855b35176aacea13e166e969307f1f27f363d30f243763cd89f5773/crisprzip-1.2.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-07-31 13:37:25",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "hiddeoff",
"github_project": "crisprzip",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "crisprzip"
}
