# Dash Cytoscape [](https://github.com/plotly/dash-cytoscape/blob/master/LICENSE) [](https://pypi.org/project/dash-cytoscape/)
<div align="center">
<a href="https://dash.plotly.com/project-maintenance">
<img src="https://dash.plotly.com/assets/images/maintained-by-plotly.png" width="400px" alt="Maintained by Plotly">
</a>
</div>
[](https://circleci.com/gh/plotly/dash-cytoscape)
A Dash component library for creating interactive and customizable networks in Python, wrapped around [Cytoscape.js](http://js.cytoscape.org/).
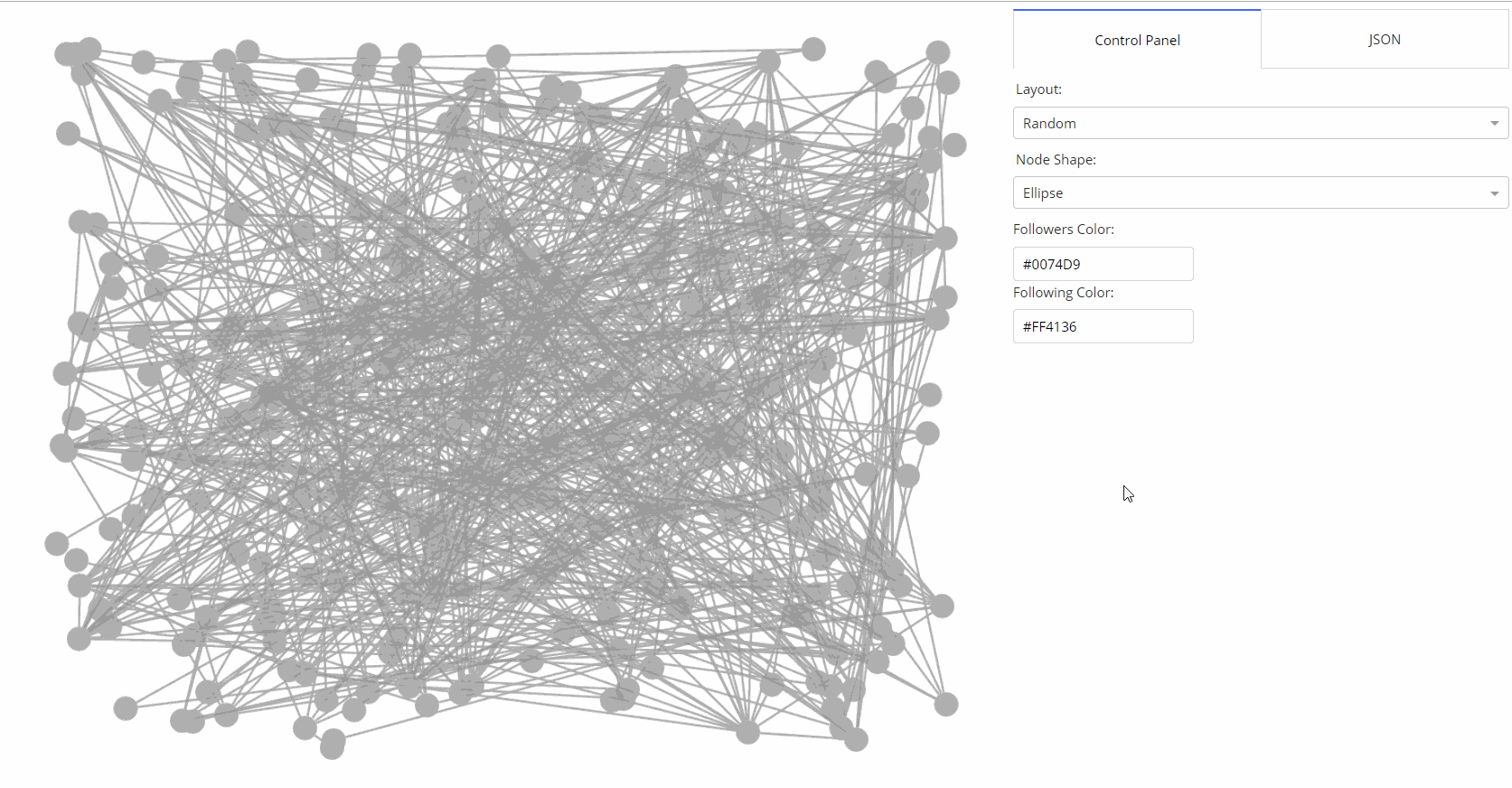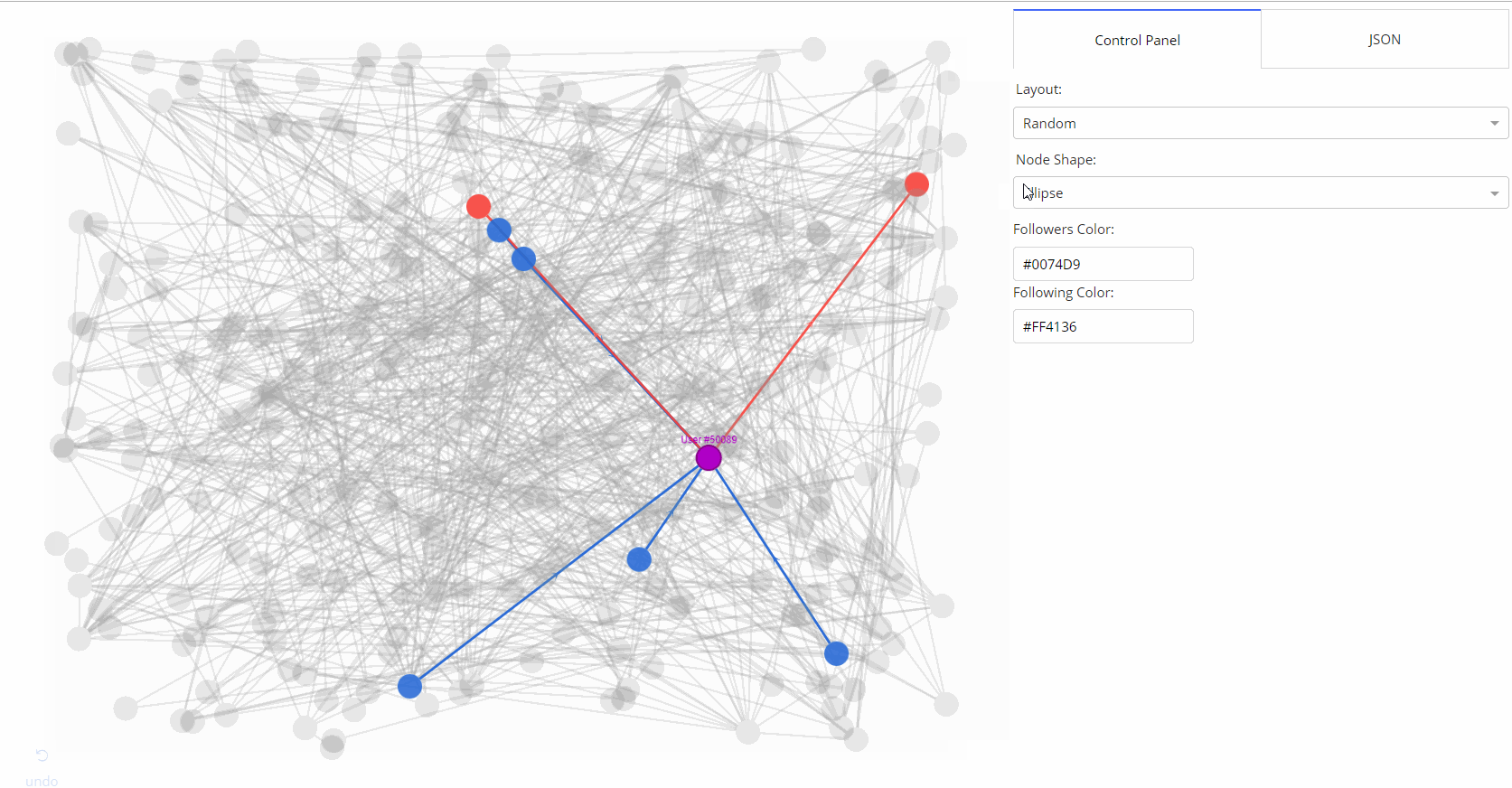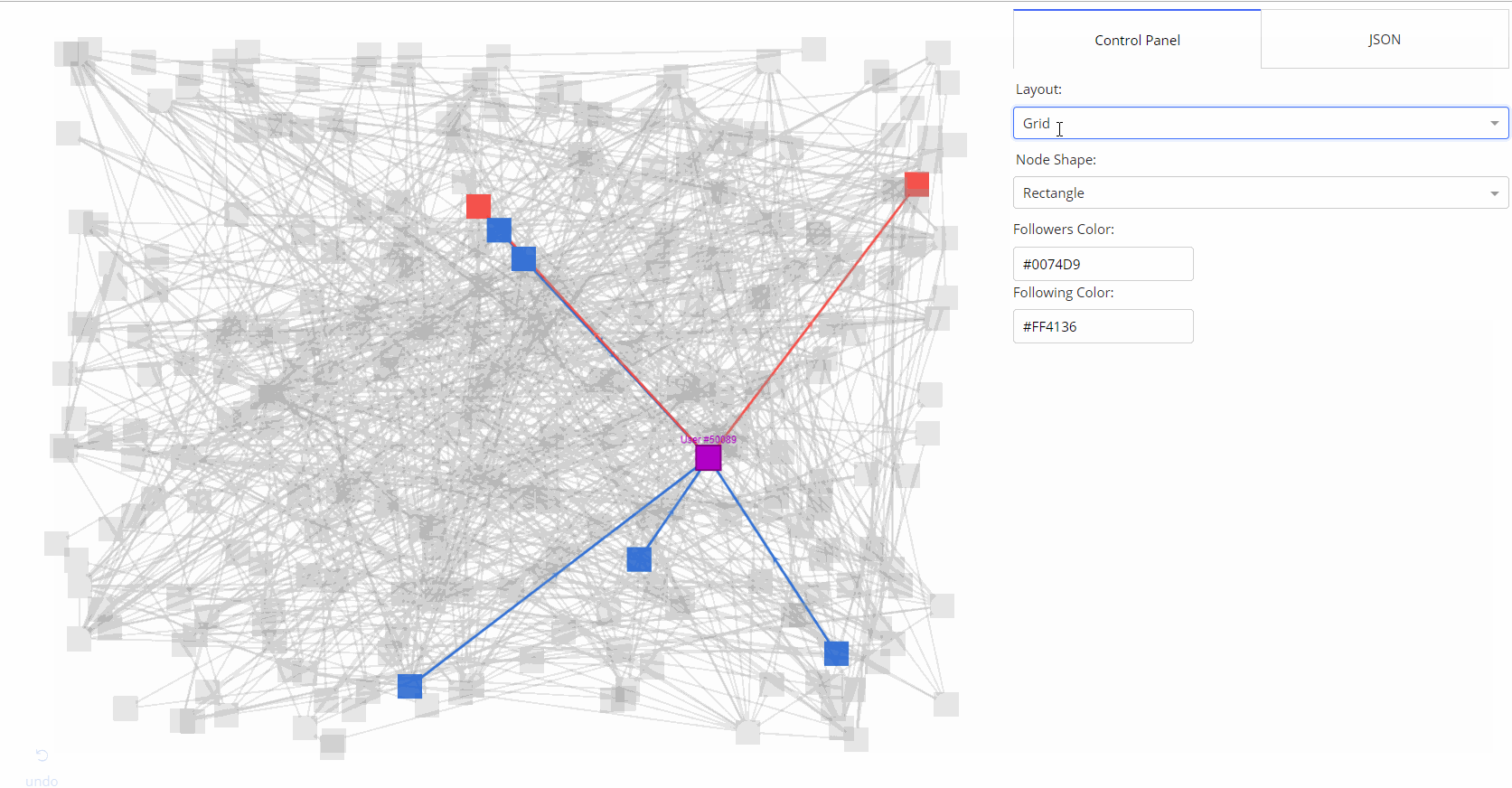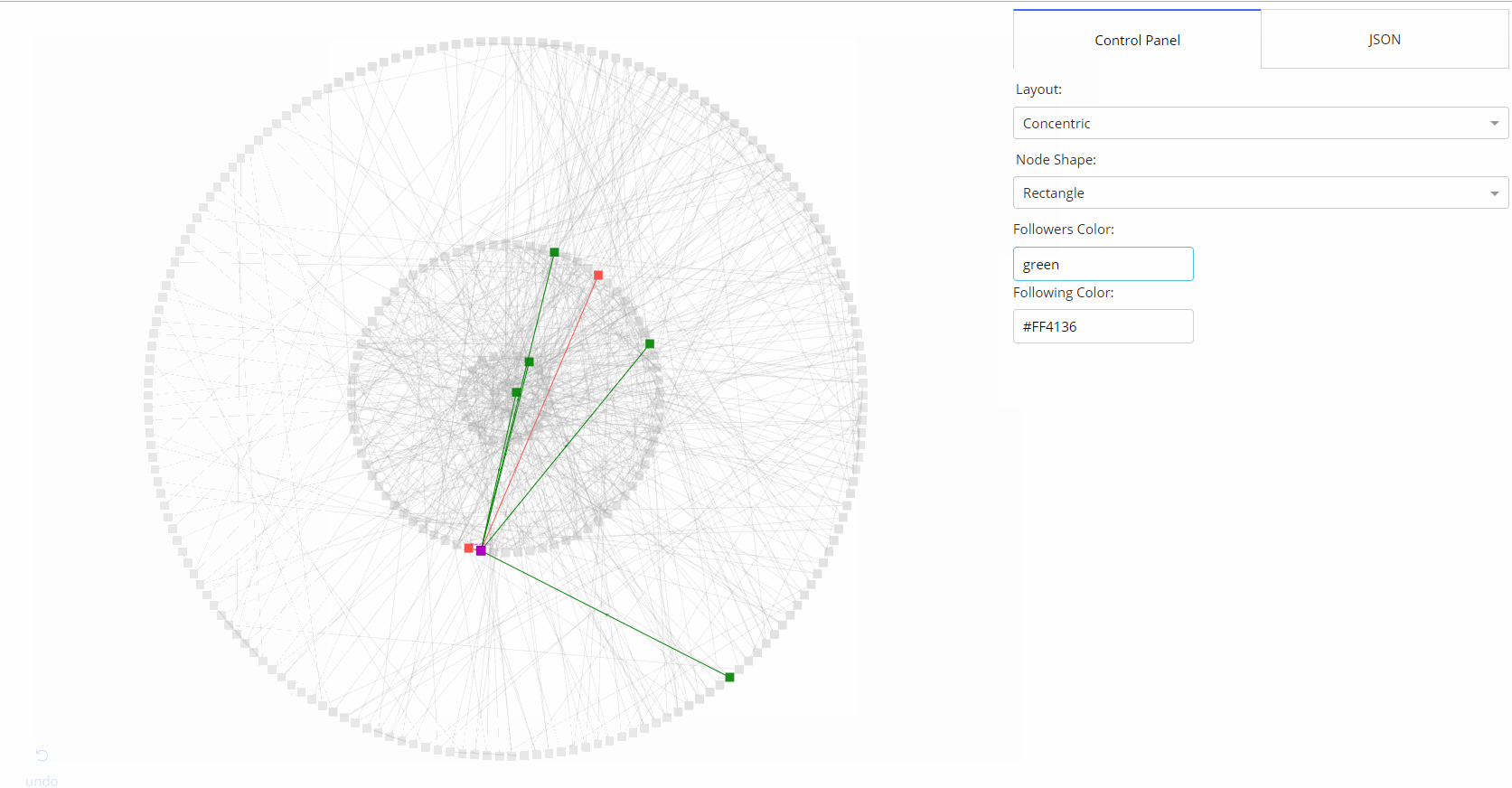
- 🌟 [Medium Article](https://medium.com/@plotlygraphs/introducing-dash-cytoscape-ce96cac824e4)
- 📣 [Community Announcement](https://community.plotly.com/t/announcing-dash-cytoscape/19095)
- 💻 [Github Repository](https://github.com/plotly/dash-cytoscape)
- 📚 [User Guide](https://dash.plotly.com/cytoscape)
- 🗺 [Component Reference](https://dash.plotly.com/cytoscape/reference)
- 📺 [Webinar Recording](https://www.youtube.com/watch?v=snXcIsCMQgk)
## Getting Started in Python
### Prerequisites
Make sure that dash and its dependent libraries are correctly installed:
```commandline
pip install dash
```
If you want to install the latest versions, check out the [Dash docs on installation](https://dash.plotly.com/installation).
### Usage
Install the library using `pip`:
```
pip install dash-cytoscape
```
If you wish to use the CyLeaflet mapping extension, you must install the optional `leaflet` dependencies:
```
pip install dash-cytoscape[leaflet]
```
Create the following example inside an `app.py` file:
```python
import dash
import dash_cytoscape as cyto
from dash import html
app = dash.Dash(__name__)
app.layout = html.Div([
cyto.Cytoscape(
id='cytoscape',
elements=[
{'data': {'id': 'one', 'label': 'Node 1'}, 'position': {'x': 50, 'y': 50}},
{'data': {'id': 'two', 'label': 'Node 2'}, 'position': {'x': 200, 'y': 200}},
{'data': {'source': 'one', 'target': 'two','label': 'Node 1 to 2'}}
],
layout={'name': 'preset'}
)
])
if __name__ == '__main__':
app.run_server(debug=True)
```
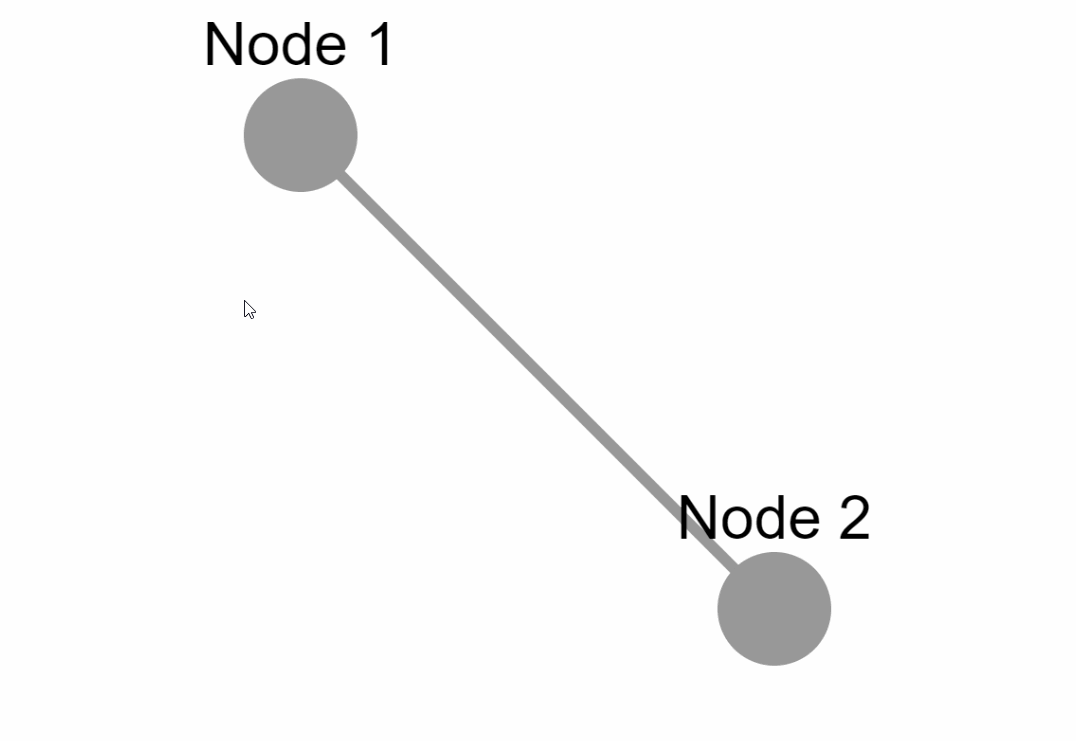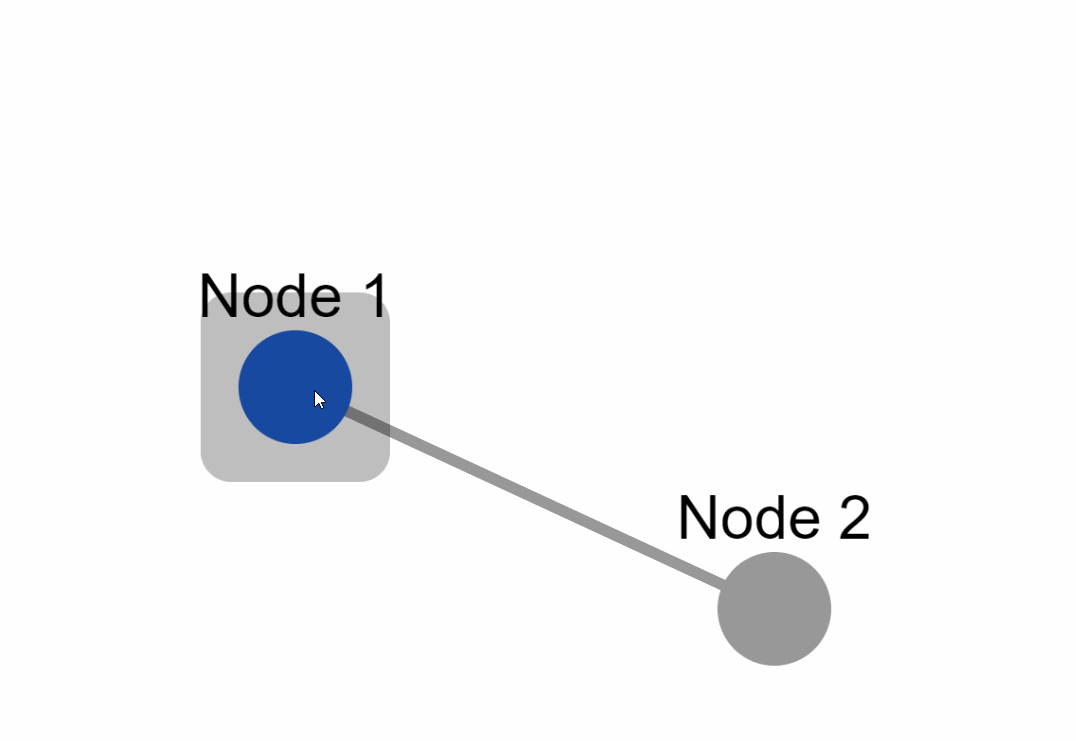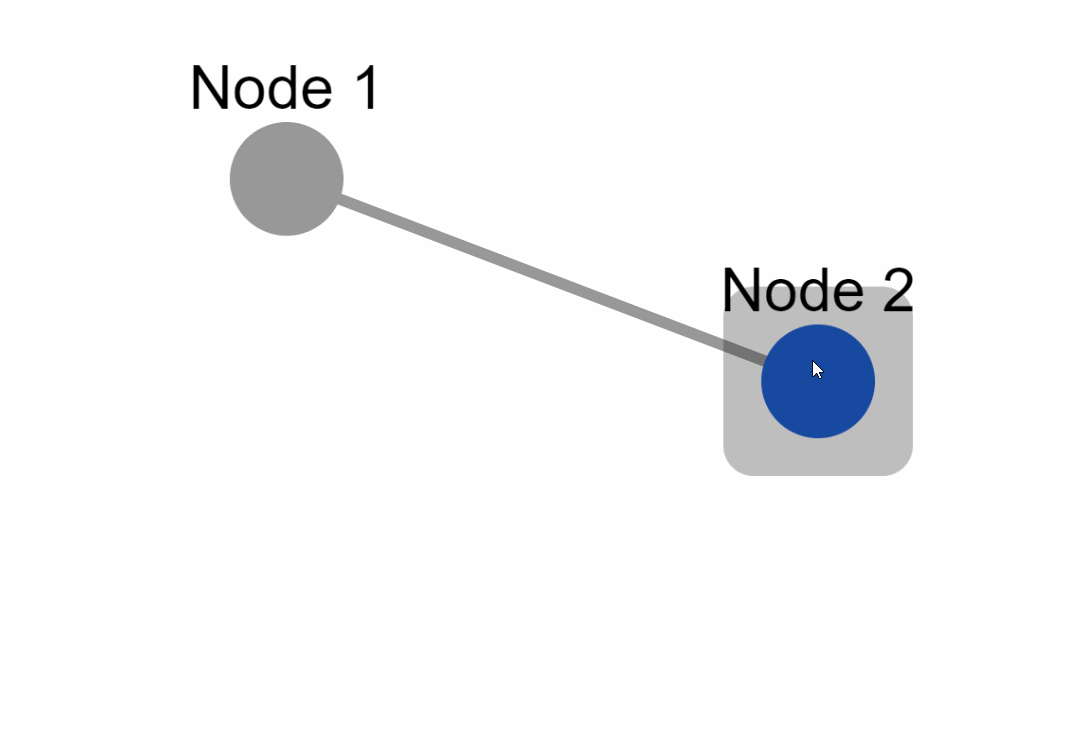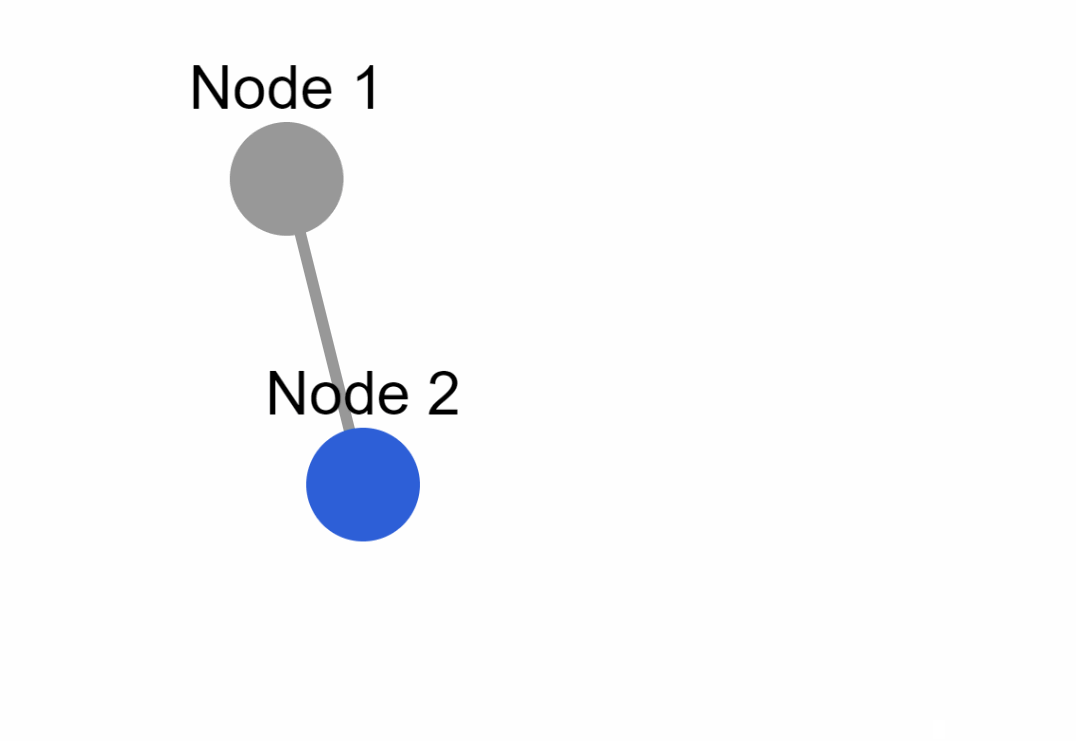
### External layouts
You can also add external layouts. Use the `cyto.load_extra_layouts()` function to get started:
```python
import dash
import dash_cytoscape as cyto
from dash import html
cyto.load_extra_layouts()
app = dash.Dash(__name__)
app.layout = html.Div([
cyto.Cytoscape(...)
])
```
Calling `cyto.load_extra_layouts()` also enables generating SVG images.
## Getting Started in R
### Prerequisites
```R
install.packages(c("devtools", "dash"))
```
### Usage
Install the library using devtools:
```
devtools::install_github("plotly/dash-cytoscape")
```
Create the following example inside an `app.R` file:
```R
library(dash)
library(dashHtmlComponents)
library(dashCytoscape)
app <- Dash$new()
app$layout(
htmlDiv(
list(
cytoCytoscape(
id = 'cytoscape-two-nodes',
layout = list('name' = 'preset'),
style = list('width' = '100%', 'height' = '400px'),
elements = list(
list('data' = list('id' = 'one', 'label' = 'Node 1'), 'position' = list('x' = 75, 'y' = 75)),
list('data' = list('id' = 'two', 'label' = 'Node 2'), 'position' = list('x' = 200, 'y' = 200)),
list('data' = list('source' = 'one', 'target' = 'two'))
)
)
)
)
)
app$run_server()
```
## Documentation
The [Dash Cytoscape User Guide](https://dash.plotly.com/cytoscape/) contains everything you need to know about the library. It contains useful examples, functioning code, and is fully interactive. You can also use the [component reference](https://dash.plotly.com/cytoscape/reference/) for a complete and concise specification of the API.
To learn more about the core Dash components and how to use callbacks, view the [Dash documentation](https://dash.plotly.com/).
For supplementary information about the underlying Javascript API, view the [Cytoscape.js documentation](http://js.cytoscape.org/).
## Contributing
Make sure that you have read and understood our [code of conduct](CODE_OF_CONDUCT.md), then head over to [CONTRIBUTING](CONTRIBUTING.md) to get started.
### Testing
Instructions on how to run [tests](CONTRIBUTING.md#tests) are given in [CONTRIBUTING.md](CONTRIBUTING.md).
## License
Dash, Cytoscape.js and Dash Cytoscape are licensed under MIT. Please view [LICENSE](LICENSE) for more details.
## Contact and Support
See https://plotly.com/dash/support for ways to get in touch.
## Acknowledgments
Huge thanks to the Cytoscape Consortium and the Cytoscape.js team for their contribution in making such a complete API for creating interactive networks. This library would not have been possible without their massive work!
The Pull Request and Issue Templates were inspired from the
[scikit-learn project](https://github.com/scikit-learn/scikit-learn).
## Gallery
### Dynamically expand elements
[Code](usage-elements.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-elements)

### Interactively update stylesheet
[Code](usage-stylesheet.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-stylesheet)

### Automatically generate interactive phylogeny trees
[Code](demos/usage-phylogeny.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-phylogeny/)

### Create your own stylesheet
[Code](usage-advanced.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-advanced)

### Use event callbacks
[Code](usage-events.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-events)

### Use external layouts
[Code](demos/usage-elements-extra.py)

### Use export graph as image
[Code](demos/usage-image-export.py)

### Make graph responsive
[Code](demos/usage-responsive-graph.py)

For an extended gallery, visit the [demos' readme](demos/README.md).
Raw data
{
"_id": null,
"home_page": "https://dash.plotly.com/cytoscape",
"name": "dash-cytoscape",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": null,
"keywords": null,
"author": "The Plotly Team <cytoscape@plotly.com>",
"author_email": "cytoscape@plotly.com",
"download_url": "https://files.pythonhosted.org/packages/ea/b7/0d511af853024241dc3192bea77e4753ea606187bd2dd777a8209a5b01bb/dash_cytoscape-1.0.2.tar.gz",
"platform": null,
"description": "# Dash Cytoscape [](https://github.com/plotly/dash-cytoscape/blob/master/LICENSE) [](https://pypi.org/project/dash-cytoscape/)\n\n<div align=\"center\">\n <a href=\"https://dash.plotly.com/project-maintenance\">\n <img src=\"https://dash.plotly.com/assets/images/maintained-by-plotly.png\" width=\"400px\" alt=\"Maintained by Plotly\">\n </a>\n</div>\n\n\n[](https://circleci.com/gh/plotly/dash-cytoscape)\n\nA Dash component library for creating interactive and customizable networks in Python, wrapped around [Cytoscape.js](http://js.cytoscape.org/).\n\n\n\n- \ud83c\udf1f [Medium Article](https://medium.com/@plotlygraphs/introducing-dash-cytoscape-ce96cac824e4)\n- \ud83d\udce3 [Community Announcement](https://community.plotly.com/t/announcing-dash-cytoscape/19095)\n- \ud83d\udcbb [Github Repository](https://github.com/plotly/dash-cytoscape)\n- \ud83d\udcda [User Guide](https://dash.plotly.com/cytoscape)\n- \ud83d\uddfa [Component Reference](https://dash.plotly.com/cytoscape/reference)\n- \ud83d\udcfa [Webinar Recording](https://www.youtube.com/watch?v=snXcIsCMQgk)\n\n## Getting Started in Python\n\n### Prerequisites\n\nMake sure that dash and its dependent libraries are correctly installed:\n\n```commandline\npip install dash\n```\n\nIf you want to install the latest versions, check out the [Dash docs on installation](https://dash.plotly.com/installation).\n\n### Usage\n\nInstall the library using `pip`:\n\n```\npip install dash-cytoscape\n```\n\nIf you wish to use the CyLeaflet mapping extension, you must install the optional `leaflet` dependencies:\n\n```\npip install dash-cytoscape[leaflet]\n```\n\nCreate the following example inside an `app.py` file:\n\n```python\nimport dash\nimport dash_cytoscape as cyto\nfrom dash import html\n\napp = dash.Dash(__name__)\napp.layout = html.Div([\n cyto.Cytoscape(\n id='cytoscape',\n elements=[\n {'data': {'id': 'one', 'label': 'Node 1'}, 'position': {'x': 50, 'y': 50}},\n {'data': {'id': 'two', 'label': 'Node 2'}, 'position': {'x': 200, 'y': 200}},\n {'data': {'source': 'one', 'target': 'two','label': 'Node 1 to 2'}}\n ],\n layout={'name': 'preset'}\n )\n])\n\nif __name__ == '__main__':\n app.run_server(debug=True)\n```\n\n\n\n### External layouts\n\nYou can also add external layouts. Use the `cyto.load_extra_layouts()` function to get started:\n\n```python\nimport dash\nimport dash_cytoscape as cyto\nfrom dash import html\n\ncyto.load_extra_layouts()\n\napp = dash.Dash(__name__)\napp.layout = html.Div([\n cyto.Cytoscape(...)\n])\n```\n\nCalling `cyto.load_extra_layouts()` also enables generating SVG images.\n\n## Getting Started in R\n\n### Prerequisites\n\n```R\ninstall.packages(c(\"devtools\", \"dash\"))\n```\n\n### Usage\n\nInstall the library using devtools:\n\n```\ndevtools::install_github(\"plotly/dash-cytoscape\")\n```\n\nCreate the following example inside an `app.R` file:\n\n```R\nlibrary(dash)\nlibrary(dashHtmlComponents)\nlibrary(dashCytoscape)\n\napp <- Dash$new()\n\napp$layout(\n htmlDiv(\n list(\n cytoCytoscape(\n id = 'cytoscape-two-nodes',\n layout = list('name' = 'preset'),\n style = list('width' = '100%', 'height' = '400px'),\n elements = list(\n list('data' = list('id' = 'one', 'label' = 'Node 1'), 'position' = list('x' = 75, 'y' = 75)),\n list('data' = list('id' = 'two', 'label' = 'Node 2'), 'position' = list('x' = 200, 'y' = 200)),\n list('data' = list('source' = 'one', 'target' = 'two'))\n )\n )\n )\n )\n)\n\napp$run_server()\n```\n\n## Documentation\n\nThe [Dash Cytoscape User Guide](https://dash.plotly.com/cytoscape/) contains everything you need to know about the library. It contains useful examples, functioning code, and is fully interactive. You can also use the [component reference](https://dash.plotly.com/cytoscape/reference/) for a complete and concise specification of the API.\n\nTo learn more about the core Dash components and how to use callbacks, view the [Dash documentation](https://dash.plotly.com/).\n\nFor supplementary information about the underlying Javascript API, view the [Cytoscape.js documentation](http://js.cytoscape.org/).\n\n## Contributing\n\nMake sure that you have read and understood our [code of conduct](CODE_OF_CONDUCT.md), then head over to [CONTRIBUTING](CONTRIBUTING.md) to get started.\n\n### Testing\n\nInstructions on how to run [tests](CONTRIBUTING.md#tests) are given in [CONTRIBUTING.md](CONTRIBUTING.md).\n\n## License\n\nDash, Cytoscape.js and Dash Cytoscape are licensed under MIT. Please view [LICENSE](LICENSE) for more details.\n\n## Contact and Support\n\nSee https://plotly.com/dash/support for ways to get in touch.\n\n## Acknowledgments\n\nHuge thanks to the Cytoscape Consortium and the Cytoscape.js team for their contribution in making such a complete API for creating interactive networks. This library would not have been possible without their massive work!\n\nThe Pull Request and Issue Templates were inspired from the\n[scikit-learn project](https://github.com/scikit-learn/scikit-learn).\n\n## Gallery\n\n### Dynamically expand elements\n\n[Code](usage-elements.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-elements)\n\n\n### Interactively update stylesheet\n\n[Code](usage-stylesheet.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-stylesheet)\n\n\n### Automatically generate interactive phylogeny trees\n\n[Code](demos/usage-phylogeny.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-phylogeny/)\n\n\n### Create your own stylesheet\n\n[Code](usage-advanced.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-advanced)\n\n\n### Use event callbacks\n\n[Code](usage-events.py) | [Demo](https://dash-gallery.plotly.host/cytoscape-events)\n\n\n### Use external layouts\n\n[Code](demos/usage-elements-extra.py)\n\n\n### Use export graph as image\n\n[Code](demos/usage-image-export.py)\n\n\n### Make graph responsive\n\n[Code](demos/usage-responsive-graph.py)\n\n\nFor an extended gallery, visit the [demos' readme](demos/README.md).\n\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A Component Library for Dash aimed at facilitating network visualization in Python, wrapped around Cytoscape.js",
"version": "1.0.2",
"project_urls": {
"Homepage": "https://dash.plotly.com/cytoscape"
},
"split_keywords": [],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "eab70d511af853024241dc3192bea77e4753ea606187bd2dd777a8209a5b01bb",
"md5": "95aae8bc551e53b116a0c8e03f67b2d5",
"sha256": "a61019d2184d63a2b3b5c06d056d3b867a04223a674cc3c7cf900a561a9a59aa"
},
"downloads": -1,
"filename": "dash_cytoscape-1.0.2.tar.gz",
"has_sig": false,
"md5_digest": "95aae8bc551e53b116a0c8e03f67b2d5",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 3992593,
"upload_time": "2024-07-15T11:39:06",
"upload_time_iso_8601": "2024-07-15T11:39:06.185699Z",
"url": "https://files.pythonhosted.org/packages/ea/b7/0d511af853024241dc3192bea77e4753ea606187bd2dd777a8209a5b01bb/dash_cytoscape-1.0.2.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-07-15 11:39:06",
"github": false,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"lcname": "dash-cytoscape"
}