## Identifying T cell antigen at the atom level with graph convolutional network
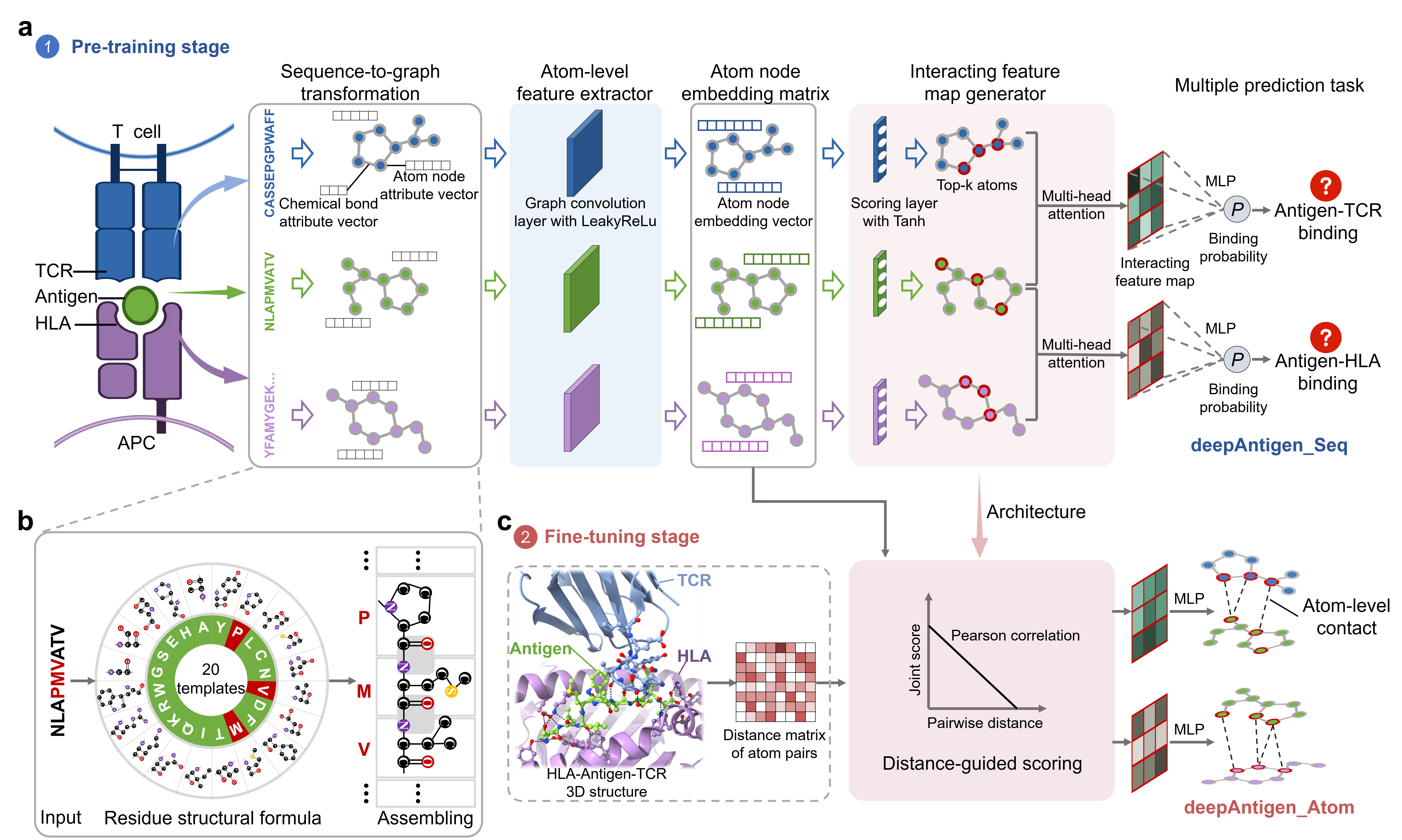
Precise identification of T cell antigen is crucial for the development of cancer mRNA vaccine. However, existing computational methods identify the interaction between antigen and Human Leukocyte Antigens (HLA) or T cell receptor (TCR) only at the sequence or residue level, which fails to capture atom-level binding patterns. In this study, we innovatively transformed each residue sequence into a topological graph, in which each node corresponds to an atom and each edge corresponds to chemical bond, and then proposed a graph convolutional network framework, called deepAntigen, to identify the interactions between antigens and TCR/HLA at the atom level. Compared to the current state-of-the-art methods, deepAntigen achieves the best performance in identifying antigens presented by HLA and recognized by TCR. Importantly, deepAntigen can discovery antigen-specific TCR motifs and capture the mutation effect on T cell immune response, facilitating to decipher underlying binding mechanism. Overall, we provide a novel method named deepAntigen for accurately identifying T cell antigens, which will contribute to the development of personalized neoantigen-targeted immunotherapies for cancer patients.
## How to install deepAntigen
To install deepAntigen, make sure you have installed [PyTorch](https://pytorch.org/) and [PyTorch Geometric](https://pytorch-geometric.readthedocs.io/en/latest/). If you need more details on the dependences, look at the environment.yml file.
- set up conda environment for deepAntigen
```shell
conda create -n deepAntigen-env python=3.8
```
- install deepAntigen from shell
```shell
pip install deepAntigen
pip install torch==1.9.0+cu111 -f https://download.pytorch.org/whl/torch_stable.html
pip install torch-cluster==1.5.9 torch-scatter==2.0.7 torch-sparse==0.6.12 torch-spline-conv==1.2.1 -f https://data.pyg.org/whl/torch-1.9.0%2Bcu111.html
pip install torch-geometric==2.4.0
```
## How to import deepAntigen
Using deepAntigen to achieve different tasks, please import corresponding module to your jupyter notebooks or scripts.
If you want to predict antigen-HLAI binding at the sequence level,
```python
from deepantigen.antigenHLAI import run_antigenHLAI_seq
```
If you want to predict atom-level contact between antigen and HLAI,
```python
from deepantigen.antigenHLAI import run_antigenHLAI_atom
```
If you want to predict antigen-HLAII binding at the sequence level,
```python
from deepantigen.antigenHLAII import run_antigenHLAII_seq
```
If you want to predict atom-level contact between antigen and HLAII,
```python
from deepantigen.antigenHLAII import run_antigenHLAII_atom
```
If you want to predict antigen-TCR binding at the sequence level,
```python
from deepantigen.antigenTCR import run_antigenTCR_seq
```
If you want to predict atom-level contact between antigen and TCR,
```python
from deepantigen.antigenTCR import run_antigenTCR_atom
```
## How to run deepAntigen for antigen-HLAI binding prediction
For sequence-level prediction, please prepare your antigen-HLAI data and place them in a .csv file format similar to the test_antigenHLAI/sequence/test.csv provided. The column 'label' is optional.
```python
df = run_antigenHLAI_seq.Inference(path)
```
The returned DataFrame, `df`, is prediction results of deepAntigen, which includes the binding probability for each antigen-HLAI pair.
For atom-level prediction, please prepare your antigen-HLAI data and place them in a .csv file format similar to the test_antigenHLAI/crystal_structure/sample.csv provided.
```python
peptide_atoms, HLAI_atoms, contact_maps = run_antigenHLAI_atom.Inference(path)
```
The returned three lists correspond top-_k_ atoms of the peptide, top-_k_ atoms of the HLAI and atom-level contact probability. Each element in `peptide_atoms` or`HLAI_atoms` is a list with length of _k_. Each element in `contact_maps` is a _k*k_ DataFrame.
If you want to train deepAntigen with your own antigen-HLAI binding data, please reference the detailed [Documentaion](#VCGRP) about deepAntigen.
## How to run deepAntigen for antigen-HLAII binding prediction
For sequence-level prediction, please prepare your antigen-HLAII data and place them in a .csv file format similar to the test_antigenHLAII/sequence/test.csv provided. The column 'label' is optional.
```python
df = run_antigenHLAII_seq.Inference(path)
```
The returned DataFrame, `df`, is prediction results of deepAntigen, which includes the binding probability for each antigen-HLAII pair.
For atom-level prediction, please prepare your antigen-HLAII data and place them in a .csv file format similar to the test_antigenHLAII/crystal_structure/sample.csv provided.
```python
peptide_atoms, HLAII_atoms, contact_maps = run_antigenHLAII_atom.Inference(path)
```
The returned three lists correspond top-_k_ atoms of the peptide, top-_k_ atoms of the HLAII and atom-level contact probability. Each element in `peptide_atoms` or `HLAII_atoms` is a list with length of _k_. Each element in `contact_maps` is a _k*k_ DataFrame.
If you want to train deepAntigen with your own antigen-HLAII binding data, please reference the detailed [Documentaion](#VCGRP) about deepAntigen.
## How to run deepAntigen for antigen-TCR binding prediction
For sequence-level prediction, please prepare your antigen-TCR data and place them in a .csv file format similar to the test_antigenTCR/sequence/test.csv provided. The column 'label' is optional.
```python
df = run_antigenTCR_seq.Inference(path)
```
The returned DataFrame, `df`, is prediction results of deepAntigen, which includes the binding probability for each antigen-TCR pair.
For atom-level prediction, please prepare your antigen-TCR data and place them in a .csv file format similar to the test_antigenTCR/crystal_structure/sample.csv provided.
```python
peptide_atoms, TCR_atoms, contact_maps = run_antigenTCR_atom.Inference(path)
```
The returned three lists correspond top-_k_ atoms of the peptide, top-_k_ atoms of the TCR and atom-level contact probability. Each element in `peptide_atoms` or `TCR_atoms` is a list with length of _k_. Each element in `contact_maps` is a _k*k_ DataFrame.
If you want to train deepAntigen with your own antigen-TCR binding data, please reference the detailed [Documentaion](#VCGRP) about deepAntigen.
## Documentation
See detailed documentation and examples at [https://deepAntigen.readthedocs.io/en/latest/index.html](https://deepAntigen.readthedocs.io/en/latest/index.html).
## Contact
Feel free to submit an issue or contact us at [quejinhao2021@163.com](mailto:quejinhao2021@163.com) for problems about the package.
Raw data
{
"_id": null,
"home_page": "https://github.com/JiangBioLab/deepAntigen",
"name": "deepAntigen",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": null,
"keywords": "T cell antigen, atom-level, GCN",
"author": "JinhaoQue",
"author_email": "quejinhao2021@163.com",
"download_url": "https://files.pythonhosted.org/packages/73/58/c59a0b8e9ca2ccdecd17f9fe4e674f0c5a14661ddcf97ece216b34a86dc0/deepantigen-1.0.3.tar.gz",
"platform": null,
"description": "## Identifying T cell antigen at the atom level with graph convolutional network\n\nPrecise identification of T cell antigen is crucial for the development of cancer mRNA vaccine. However, existing computational methods identify the interaction between antigen and Human Leukocyte Antigens (HLA) or T cell receptor (TCR) only at the sequence or residue level, which fails to capture atom-level binding patterns. In this study, we innovatively transformed each residue sequence into a topological graph, in which each node corresponds to an atom and each edge corresponds to chemical bond, and then proposed a graph convolutional network framework, called deepAntigen, to identify the interactions between antigens and TCR/HLA at the atom level. Compared to the current state-of-the-art methods, deepAntigen achieves the best performance in identifying antigens presented by HLA and recognized by TCR. Importantly, deepAntigen can discovery antigen-specific TCR motifs and capture the mutation effect on T cell immune response, facilitating to decipher underlying binding mechanism. Overall, we provide a novel method named deepAntigen for accurately identifying T cell antigens, which will contribute to the development of personalized neoantigen-targeted immunotherapies for cancer patients.\n## How to install deepAntigen\nTo install deepAntigen, make sure you have installed [PyTorch](https://pytorch.org/) and [PyTorch Geometric](https://pytorch-geometric.readthedocs.io/en/latest/). If you need more details on the dependences, look at the environment.yml file.\n\n- set up conda environment for deepAntigen\n```shell\nconda create -n deepAntigen-env python=3.8\n```\n\n- install deepAntigen from shell\n```shell\npip install deepAntigen\npip install torch==1.9.0+cu111 -f https://download.pytorch.org/whl/torch_stable.html\npip install torch-cluster==1.5.9 torch-scatter==2.0.7 torch-sparse==0.6.12 torch-spline-conv==1.2.1 -f https://data.pyg.org/whl/torch-1.9.0%2Bcu111.html\npip install torch-geometric==2.4.0\n```\n\n## How to import deepAntigen\nUsing deepAntigen to achieve different tasks, please import corresponding module to your jupyter notebooks or scripts. \n\nIf you want to predict antigen-HLAI binding at the sequence level, \n```python\nfrom deepantigen.antigenHLAI import run_antigenHLAI_seq\n```\nIf you want to predict atom-level contact between antigen and HLAI, \n```python\nfrom deepantigen.antigenHLAI import run_antigenHLAI_atom\n```\n\nIf you want to predict antigen-HLAII binding at the sequence level, \n```python\nfrom deepantigen.antigenHLAII import run_antigenHLAII_seq\n```\nIf you want to predict atom-level contact between antigen and HLAII, \n```python\nfrom deepantigen.antigenHLAII import run_antigenHLAII_atom\n```\n\nIf you want to predict antigen-TCR binding at the sequence level, \n```python\nfrom deepantigen.antigenTCR import run_antigenTCR_seq\n```\nIf you want to predict atom-level contact between antigen and TCR,\n```python\nfrom deepantigen.antigenTCR import run_antigenTCR_atom\n```\n## How to run deepAntigen for antigen-HLAI binding prediction\nFor sequence-level prediction, please prepare your antigen-HLAI data and place them in a .csv file format similar to the test_antigenHLAI/sequence/test.csv provided. The column 'label' is optional. \n```python\ndf = run_antigenHLAI_seq.Inference(path)\n```\nThe returned DataFrame, `df`, is prediction results of deepAntigen, which includes the binding probability for each antigen-HLAI pair. \nFor atom-level prediction, please prepare your antigen-HLAI data and place them in a .csv file format similar to the test_antigenHLAI/crystal_structure/sample.csv provided.\n```python\npeptide_atoms, HLAI_atoms, contact_maps = run_antigenHLAI_atom.Inference(path)\n```\nThe returned three lists correspond top-_k_ atoms of the peptide, top-_k_ atoms of the HLAI and atom-level contact probability. Each element in `peptide_atoms` or`HLAI_atoms` is a list with length of _k_. Each element in `contact_maps` is a _k*k_ DataFrame.\n\nIf you want to train deepAntigen with your own antigen-HLAI binding data, please reference the detailed [Documentaion](#VCGRP) about deepAntigen.\n## How to run deepAntigen for antigen-HLAII binding prediction\nFor sequence-level prediction, please prepare your antigen-HLAII data and place them in a .csv file format similar to the test_antigenHLAII/sequence/test.csv provided. The column 'label' is optional. \n```python\ndf = run_antigenHLAII_seq.Inference(path)\n```\nThe returned DataFrame, `df`, is prediction results of deepAntigen, which includes the binding probability for each antigen-HLAII pair. \nFor atom-level prediction, please prepare your antigen-HLAII data and place them in a .csv file format similar to the test_antigenHLAII/crystal_structure/sample.csv provided.\n```python\npeptide_atoms, HLAII_atoms, contact_maps = run_antigenHLAII_atom.Inference(path)\n```\nThe returned three lists correspond top-_k_ atoms of the peptide, top-_k_ atoms of the HLAII and atom-level contact probability. Each element in `peptide_atoms` or `HLAII_atoms` is a list with length of _k_. Each element in `contact_maps` is a _k*k_ DataFrame.\n\nIf you want to train deepAntigen with your own antigen-HLAII binding data, please reference the detailed [Documentaion](#VCGRP) about deepAntigen.\n## How to run deepAntigen for antigen-TCR binding prediction\nFor sequence-level prediction, please prepare your antigen-TCR data and place them in a .csv file format similar to the test_antigenTCR/sequence/test.csv provided. The column 'label' is optional. \n```python\ndf = run_antigenTCR_seq.Inference(path)\n```\nThe returned DataFrame, `df`, is prediction results of deepAntigen, which includes the binding probability for each antigen-TCR pair. \nFor atom-level prediction, please prepare your antigen-TCR data and place them in a .csv file format similar to the test_antigenTCR/crystal_structure/sample.csv provided.\n```python\npeptide_atoms, TCR_atoms, contact_maps = run_antigenTCR_atom.Inference(path)\n```\nThe returned three lists correspond top-_k_ atoms of the peptide, top-_k_ atoms of the TCR and atom-level contact probability. Each element in `peptide_atoms` or `TCR_atoms` is a list with length of _k_. Each element in `contact_maps` is a _k*k_ DataFrame.\n\nIf you want to train deepAntigen with your own antigen-TCR binding data, please reference the detailed [Documentaion](#VCGRP) about deepAntigen.\n## Documentation\nSee detailed documentation and examples at [https://deepAntigen.readthedocs.io/en/latest/index.html](https://deepAntigen.readthedocs.io/en/latest/index.html).\n## Contact\nFeel free to submit an issue or contact us at [quejinhao2021@163.com](mailto:quejinhao2021@163.com) for problems about the package.\n",
"bugtrack_url": null,
"license": "MIT License",
"summary": "Identifying T cell antigen at the atomic level with graph convolutional network",
"version": "1.0.3",
"project_urls": {
"Homepage": "https://github.com/JiangBioLab/deepAntigen"
},
"split_keywords": [
"t cell antigen",
" atom-level",
" gcn"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "1daf10dbb3127db0e61d84ec2210a607a2d7a015b0f48ff1fc417049e7ba1676",
"md5": "5ab3f4543e31bbee4ad468977d686349",
"sha256": "392dd84d231c453656bef69b261d6fb6c439aa7b83421c72a9943da4c33a52b8"
},
"downloads": -1,
"filename": "deepAntigen-1.0.3-py3-none-any.whl",
"has_sig": false,
"md5_digest": "5ab3f4543e31bbee4ad468977d686349",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 61623162,
"upload_time": "2025-01-30T02:44:10",
"upload_time_iso_8601": "2025-01-30T02:44:10.063823Z",
"url": "https://files.pythonhosted.org/packages/1d/af/10dbb3127db0e61d84ec2210a607a2d7a015b0f48ff1fc417049e7ba1676/deepAntigen-1.0.3-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "7358c59a0b8e9ca2ccdecd17f9fe4e674f0c5a14661ddcf97ece216b34a86dc0",
"md5": "95eebf687bb06684cf20f1d9bd616cf6",
"sha256": "4a2a311d4a875a4bc4f64c5bc114e8959df7732d0b408664598971cb136da98c"
},
"downloads": -1,
"filename": "deepantigen-1.0.3.tar.gz",
"has_sig": false,
"md5_digest": "95eebf687bb06684cf20f1d9bd616cf6",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 61590243,
"upload_time": "2025-01-30T02:41:37",
"upload_time_iso_8601": "2025-01-30T02:41:37.893310Z",
"url": "https://files.pythonhosted.org/packages/73/58/c59a0b8e9ca2ccdecd17f9fe4e674f0c5a14661ddcf97ece216b34a86dc0/deepantigen-1.0.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-01-30 02:41:37",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "JiangBioLab",
"github_project": "deepAntigen",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "deepantigen"
}
