# digitalTwinOnFHIR
## Usage
- Setup and connect to FHIR server
```python
from digitaltwin_on_fhir.core import Adapter
adapter = Adapter("http://localhost:8080/fhir/")
```
### Load data to FHIR server
#### Primary measurements
- Load FHIR bundle
```python
await adapter.loader().load_fhir_bundle('./dataset/dataset-fhir-bundles')
```
- Load DigitalTWIN Clinical Description (primary measurements)
```python
measurements = adapter.loader().load_sparc_dataset_primary_measurements()
with open('./dataset/measurements.json', 'r') as file:
data = json.load(file)
await measurements.add_measurements_description(data).generate_resources()
```
- Add Practitioner (researcher) to FHIR server
```python
from digitaltwin_on_fhir.core.resource import Identifier, Code, HumanName, Practitioner
await measurements.add_practitioner(researcher=Practitioner(
active=True,
identifier=[
Identifier(use=Code("official"), system="sparc.org",
value='sparc-d557ac68-f365-0718-c945-8722ec')],
name=[HumanName(use="usual", text="Xiaoming Li", family="Li", given=["Xiaoming"])],
gender="male"
))
```
#### Workflow
### Search
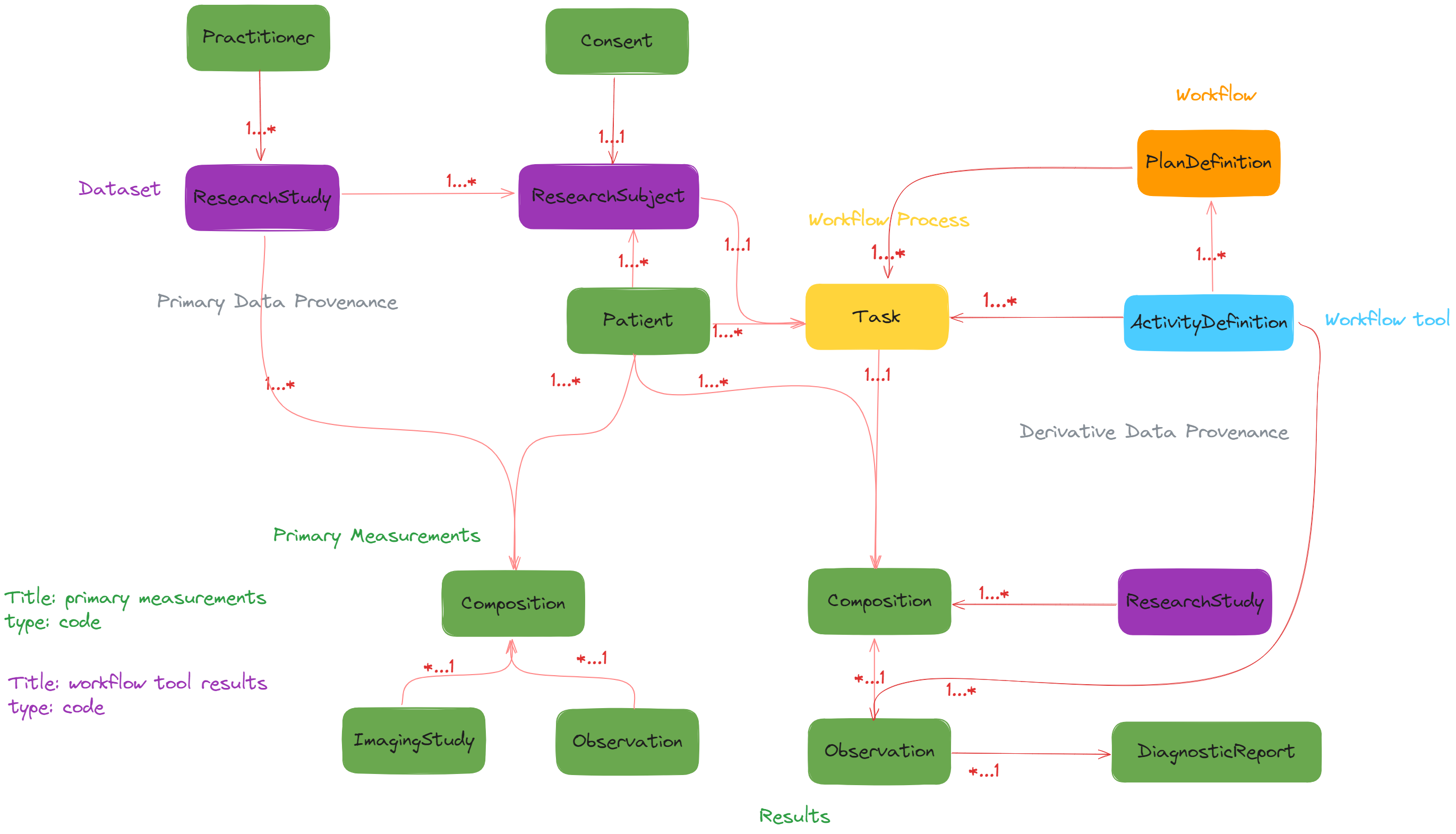
#### References in Task (workflow tool process) resource
- owner: `Patient` reference
- for: `PlanDefinition` (workflow) reference
- focus: `ActivityDefinition` (workflow tool) reference
- basedOn: `ResearchSubject` reference
- requester (Optional): `Practitioner` (researcher) reference
- references in input
- ImagingStudy
- Observation
- references in output
- Observation
###### Example
- Find a specific workflow process
- If known: patient, dataset, workflow tool and workflow uuids
```python
client = adapter.async_client
# Step 1: find the patient
patient = await client.resources("Patient").search(
identifier="patient-xxxx").first()
# Step 2: find the dataset
dataset = await client.resources("ResearchStudy").search(
identifier="dataset-xxxx").first()
# Step 3: find the workflow tool
workflow_tool = await client.resources("ActivityDefinition").search(
identifier="workflow-tool-xxxx").first()
# Step 4: find the research subject
research_subject = await client.resources("ResearchSubject").search(
patient=patient.to_reference().reference,
study=dataset.to_reference().reference).first()
# Step 5: find workflow
workflow = await client.resources("PlanDefinition").search(
identifier="sparc-workflow-uuid-001").first()
workflow_tool_process = await client.resources("Task").search(
subject=workflow.to_reference(),
focus=workflow_tool.to_reference(),
based_on=research_subject.to_reference(),
owner=patient.to_reference()).first()
```
- Find all input resources of the workflow tool process
```python
inputs = workflow_tool_process.get("input")
for i in inputs:
input_reference = i.get("valueReference")
input_resource = await input_reference.to_resource()
```
- Find the input data comes from with dataset
- Assume we don't know the dataset and patient uuids at this stage
```python
composition = await client.resources("Composition").search(
title="primary measurements",
entry=input_reference).first()
dataset = await composition.get("subject").to_resource()
```
- Find all output resources of the workflow tool process
```python
outputs = workflow_tool_process.get("output")
for output in outputs:
output_reference = output.get("valueReference")
output_resource = await output_reference.to_resource()
```
#### References in PlanDefinition (workflow) resource
- action
- definition_canonical: ActivityDefinition (workflow tool) reference
###### Example
- If known workflow uuid
- Find all related workflow tools
```python
workflow = await client.resources("PlanDefinition").search(
identifier="sparc-workflow-uuid-001").first()
actions = workflow.get("action")
for a in actions:
if a.get("definitionCanonical") is None:
continue
resource_type, _id = a.get("definitionCanonical").split("/")
workflow_tool = await client.reference(resource_type, _id).to_resource()
```
- Find all related workflow processes
```python
workflow_tool_processes = await client.resources("Task").search(
subject=workflow.to_reference()).fetch_all()
```
## DigitalTWIN on FHIR Diagram
Raw data
{
"_id": null,
"home_page": null,
"name": "digitaltwin-on-fhir",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.9",
"maintainer_email": null,
"keywords": "fhir, DigitalTWIN, SPARC, Clinic Description",
"author": null,
"author_email": "Linkun Gao <gaolinkun123@gmail.com>",
"download_url": "https://files.pythonhosted.org/packages/53/25/d2a24fda18068570a65b168bb26337dd5a94d7b753cf77931508178bf436/digitaltwin_on_fhir-1.1.2.tar.gz",
"platform": null,
"description": "# digitalTwinOnFHIR\n\n## Usage\n\n- Setup and connect to FHIR server\n\n```python\nfrom digitaltwin_on_fhir.core import Adapter\n\nadapter = Adapter(\"http://localhost:8080/fhir/\")\n```\n\n### Load data to FHIR server\n\n#### Primary measurements\n\n- Load FHIR bundle\n```python\n await adapter.loader().load_fhir_bundle('./dataset/dataset-fhir-bundles')\n```\n- Load DigitalTWIN Clinical Description (primary measurements)\n```python\nmeasurements = adapter.loader().load_sparc_dataset_primary_measurements()\nwith open('./dataset/measurements.json', 'r') as file:\n data = json.load(file)\n\nawait measurements.add_measurements_description(data).generate_resources()\n```\n- Add Practitioner (researcher) to FHIR server\n\n```python\nfrom digitaltwin_on_fhir.core.resource import Identifier, Code, HumanName, Practitioner\n\nawait measurements.add_practitioner(researcher=Practitioner(\n active=True,\n identifier=[\n Identifier(use=Code(\"official\"), system=\"sparc.org\",\n value='sparc-d557ac68-f365-0718-c945-8722ec')],\n name=[HumanName(use=\"usual\", text=\"Xiaoming Li\", family=\"Li\", given=[\"Xiaoming\"])],\n gender=\"male\"\n))\n```\n\n#### Workflow\n\n### Search\n#### References in Task (workflow tool process) resource\n- owner: `Patient` reference\n- for: `PlanDefinition` (workflow) reference\n- focus: `ActivityDefinition` (workflow tool) reference\n- basedOn: `ResearchSubject` reference\n- requester (Optional): `Practitioner` (researcher) reference\n- references in input\n - ImagingStudy\n - Observation\n- references in output\n - Observation\n\n###### Example\n\n- Find a specific workflow process\n - If known: patient, dataset, workflow tool and workflow uuids\n\n```python\nclient = adapter.async_client\n\n# Step 1: find the patient\npatient = await client.resources(\"Patient\").search(\n identifier=\"patient-xxxx\").first()\n# Step 2: find the dataset\ndataset = await client.resources(\"ResearchStudy\").search(\n identifier=\"dataset-xxxx\").first()\n# Step 3: find the workflow tool\nworkflow_tool = await client.resources(\"ActivityDefinition\").search(\n identifier=\"workflow-tool-xxxx\").first()\n# Step 4: find the research subject\nresearch_subject = await client.resources(\"ResearchSubject\").search(\n patient=patient.to_reference().reference,\n study=dataset.to_reference().reference).first()\n# Step 5: find workflow\nworkflow = await client.resources(\"PlanDefinition\").search(\n identifier=\"sparc-workflow-uuid-001\").first()\nworkflow_tool_process = await client.resources(\"Task\").search(\n subject=workflow.to_reference(),\n focus=workflow_tool.to_reference(),\n based_on=research_subject.to_reference(),\n owner=patient.to_reference()).first()\n```\n- Find all input resources of the workflow tool process\n```python\ninputs = workflow_tool_process.get(\"input\")\nfor i in inputs:\n input_reference = i.get(\"valueReference\")\n input_resource = await input_reference.to_resource()\n```\n- Find the input data comes from with dataset\n - Assume we don't know the dataset and patient uuids at this stage\n```python\ncomposition = await client.resources(\"Composition\").search(\n title=\"primary measurements\", \n entry=input_reference).first()\ndataset = await composition.get(\"subject\").to_resource()\n```\n\n- Find all output resources of the workflow tool process\n```python\noutputs = workflow_tool_process.get(\"output\")\nfor output in outputs:\n output_reference = output.get(\"valueReference\")\n output_resource = await output_reference.to_resource()\n```\n\n#### References in PlanDefinition (workflow) resource\n- action\n - definition_canonical: ActivityDefinition (workflow tool) reference\n\n###### Example\n- If known workflow uuid\n - Find all related workflow tools\n ```python\n workflow = await client.resources(\"PlanDefinition\").search(\n identifier=\"sparc-workflow-uuid-001\").first()\n actions = workflow.get(\"action\")\n \n for a in actions:\n if a.get(\"definitionCanonical\") is None:\n continue\n resource_type, _id = a.get(\"definitionCanonical\").split(\"/\")\n workflow_tool = await client.reference(resource_type, _id).to_resource()\n ```\n - Find all related workflow processes\n ```python\n workflow_tool_processes = await client.resources(\"Task\").search(\n subject=workflow.to_reference()).fetch_all()\n ```\n\n## DigitalTWIN on FHIR Diagram\n\n",
"bugtrack_url": null,
"license": null,
"summary": "An adapter for transfer DigitalTWIN Clinic Description to FHIR",
"version": "1.1.2",
"project_urls": {
"Documentation": "https://github.com/Copper3D-brids/digitalTwinOnFHIR#readme",
"Homepage": "https://github.com/Copper3D-brids/digitalTwinOnFHIR",
"Source": "https://github.com/Copper3D-brids/digitalTwinOnFHIR.git"
},
"split_keywords": [
"fhir",
" digitaltwin",
" sparc",
" clinic description"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "2b78958ad1e0ad359d9667989098eaac5042305212f97b86f034abc01cb93904",
"md5": "e3ea3d5b1e6c05cc83f19a66b7d398ce",
"sha256": "de18b71d974b4f10e212959cfde2c8880abbfd0c2893d622891b627f10f84f51"
},
"downloads": -1,
"filename": "digitaltwin_on_fhir-1.1.2-py3-none-any.whl",
"has_sig": false,
"md5_digest": "e3ea3d5b1e6c05cc83f19a66b7d398ce",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.9",
"size": 81493,
"upload_time": "2024-08-28T03:25:20",
"upload_time_iso_8601": "2024-08-28T03:25:20.232702Z",
"url": "https://files.pythonhosted.org/packages/2b/78/958ad1e0ad359d9667989098eaac5042305212f97b86f034abc01cb93904/digitaltwin_on_fhir-1.1.2-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "5325d2a24fda18068570a65b168bb26337dd5a94d7b753cf77931508178bf436",
"md5": "859797a3ed6f88eab572cbc969fde56d",
"sha256": "246ad36797ac8b3bb3e0dbc3dbd8fbd55897d49a99b9a9eea41cd61e1c1b42d7"
},
"downloads": -1,
"filename": "digitaltwin_on_fhir-1.1.2.tar.gz",
"has_sig": false,
"md5_digest": "859797a3ed6f88eab572cbc969fde56d",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.9",
"size": 63640,
"upload_time": "2024-08-28T03:25:22",
"upload_time_iso_8601": "2024-08-28T03:25:22.016831Z",
"url": "https://files.pythonhosted.org/packages/53/25/d2a24fda18068570a65b168bb26337dd5a94d7b753cf77931508178bf436/digitaltwin_on_fhir-1.1.2.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-08-28 03:25:22",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "Copper3D-brids",
"github_project": "digitalTwinOnFHIR#readme",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [],
"lcname": "digitaltwin-on-fhir"
}
