# CS410_FlyEM
This repository hosts the segmentation pipeline to segment fly retina images using traditional segmentation algorithms.
## Authors
* [Prof. Daniel Haehn](https://github.com/haehn)
* [Sai Harshavardhan Reddy Kona](https://github.com/kshvr16)
* [Nikhila Yadav Lankela](https://github.com/Nikhila1003)
* [Varshitha Hantur Dinakar](https://github.com/varshi-123)
* [Varuni Manjunath](https://github.com/Varunii)
* [Kiran Sandilya](https://github.com/Kiransandilya)
* [Kunal Jain](https://github.com/jainkhere)
## Package Installation
Install this python package by executing the following command.
```bash
pip install flyem_segmentation_pipeline
```
If the above command throws any error, please run the following command and install the dependency packages individually.
```bash
pip install flyem_segmentation_pipeline --no-deps
```
## Required Python packages
* [numpy](https://pypi.org/project/numpy/)
* [mahotas](https://pypi.org/project/mahotas/)
* [matplotlib](https://pypi.org/project/matplotlib/)
* [scikit-image](https://pypi.org/project/scikit-image/)
## Dependency package issues
All the above-mentioned packages are required to use this package, but due to the version issues with numpy and mahotas, during installation only matplotlib and scikit-image are installed; numpy and mahotas has to be installed separately.
## Package Description
Anyone can use this package to perform various computer vision algorithms to segment the fly retina images.
The package followed MVC(Model-View-Controller) design pattern.
All the functionalities can be used by importing them using the following command.
```python
from segmentationMVC.controller import ImageController
```
## Functions
**ImageController.read():** reads an image from the given path.
**ImageController.normalize():** returns an normalized image of the input image to [0, 255].
**ImageController.center_crop():** crops an image to a default size of (512, 512).
**ImageController.smooth():** smoothens the image using Gaussian filter.
**ImageController.threshold():** applies thresholding of an input image.
**ImageController.binary_mask():** creates an binary mask of the image.
**ImageController.display():** display the image.
## Sample Usage
The below sample code reads an image from the input path.
```python
from segmentationMVC.controller import ImageController
image_data = ImageController.read("..")
```
Output:
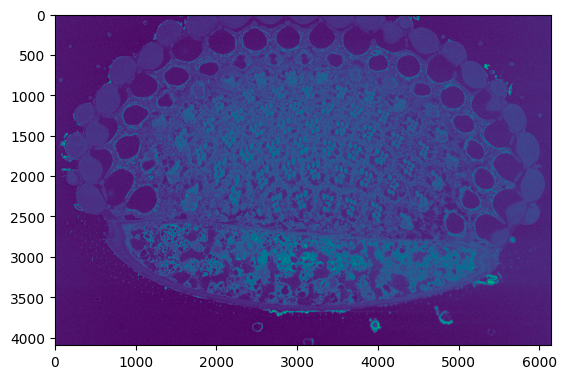
The below sample code normalized the input image.
```python
from segmentationMVC.controller import ImageController
normalized_data = ImageController.normalize(image_data)
```
Output:
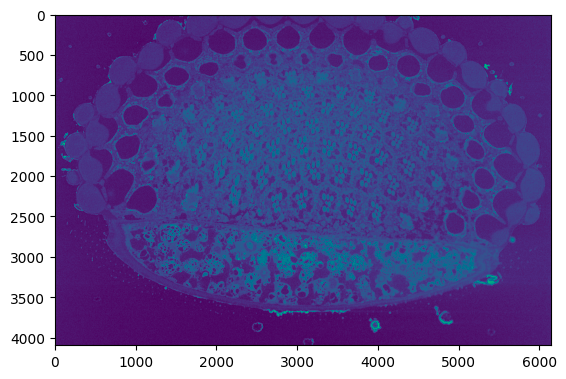
The below sample code crops the input image.
```python
from segmentationMVC.controller import ImageController
crop_data = ImageController.normalize(normalized_data)
```
Output:
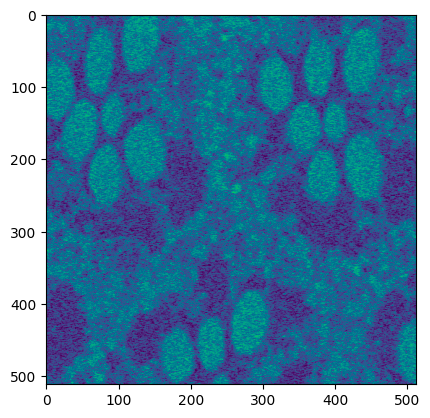
The below sample code smoothens the input image.
```python
from segmentationMVC.controller import ImageController
smoothed_data = ImageController.smooth(crop_data)
```
Output:
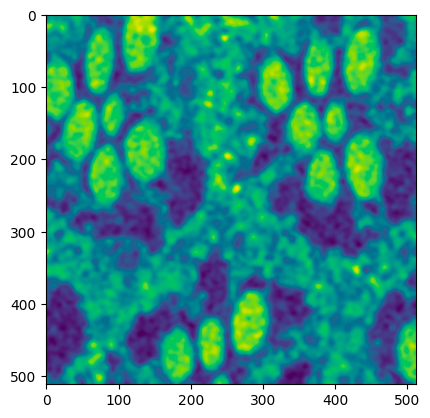
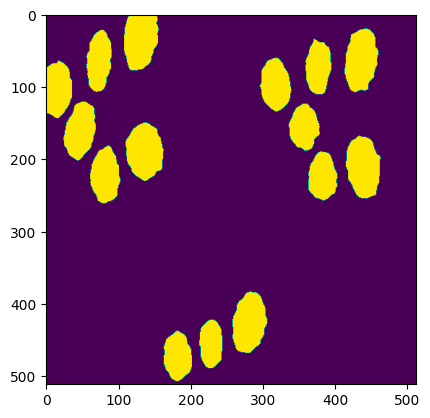
To get the complete segmented image, the following will serve as an example.
```python
from segmentationMVC.controller import ImageController
image_data = ImageController.read("..")
normalized_data = ImageController.normalize(image_data)
crop_data = ImageController.normalize(normalized_data)
smoothed_data = ImageController.smooth(crop_data)
threshold_data = ImageController.threshold(smoothed_data, threshold_value=73)
labeled_data, nr_count = ImageController.label(threshold_data)
selected_labeled_data = ImageController.select_regions(labeled_data, region_size=1500)
binary_mask_data = ImageController.binary_mask(selected_labeled_data)
closed_binary_mask_data = ImageController.close_binary_mask(binary_mask_data)
segmented_image = ImageController.binary_image(closed_binary_mask_data)
```
The variable **segmented_image** in the above sample code stores the result of the segmented image.
Output:
Raw data
{
"_id": null,
"home_page": "https://github.com/kshvr16/CS410_FlyEM_PyPi",
"name": "flyem-segmentation-pipeline",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "medical image,medical image segmentation,fly retina images,electron microscopy images",
"author": "Sai Harshavardhan Reddy Kona",
"author_email": "s.kona001@umb.edu",
"download_url": "https://files.pythonhosted.org/packages/81/6e/308b677d1cc868649b8a298aeeb94559586c924f768e01c49c050b9da066/flyem_segmentation_pipeline-1.3.3.tar.gz",
"platform": null,
"description": "# CS410_FlyEM\nThis repository hosts the segmentation pipeline to segment fly retina images using traditional segmentation algorithms.\n\n\n## Authors\n\n* [Prof. Daniel Haehn](https://github.com/haehn)\n* [Sai Harshavardhan Reddy Kona](https://github.com/kshvr16)\n* [Nikhila Yadav Lankela](https://github.com/Nikhila1003)\n* [Varshitha Hantur Dinakar](https://github.com/varshi-123)\n* [Varuni Manjunath](https://github.com/Varunii)\n* [Kiran Sandilya](https://github.com/Kiransandilya)\n* [Kunal Jain](https://github.com/jainkhere)\n\n\n## Package Installation\n\nInstall this python package by executing the following command.\n```bash\n pip install flyem_segmentation_pipeline\n```\nIf the above command throws any error, please run the following command and install the dependency packages individually.\n```bash\npip install flyem_segmentation_pipeline --no-deps\n```\n\n\n## Required Python packages\n\n* [numpy](https://pypi.org/project/numpy/)\n* [mahotas](https://pypi.org/project/mahotas/)\n* [matplotlib](https://pypi.org/project/matplotlib/)\n* [scikit-image](https://pypi.org/project/scikit-image/)\n\n ## Dependency package issues\nAll the above-mentioned packages are required to use this package, but due to the version issues with numpy and mahotas, during installation only matplotlib and scikit-image are installed; numpy and mahotas has to be installed separately.\n\n\n## Package Description\nAnyone can use this package to perform various computer vision algorithms to segment the fly retina images.\n\nThe package followed MVC(Model-View-Controller) design pattern.\n\nAll the functionalities can be used by importing them using the following command.\n```python\nfrom segmentationMVC.controller import ImageController\n```\n\n## Functions\n\n**ImageController.read():** reads an image from the given path.\n\n**ImageController.normalize():** returns an normalized image of the input image to [0, 255].\n\n**ImageController.center_crop():** crops an image to a default size of (512, 512).\n\n**ImageController.smooth():** smoothens the image using Gaussian filter.\n\n**ImageController.threshold():** applies thresholding of an input image.\n\n**ImageController.binary_mask():** creates an binary mask of the image.\n\n**ImageController.display():** display the image.\n\n## Sample Usage\nThe below sample code reads an image from the input path. \n```python\nfrom segmentationMVC.controller import ImageController\nimage_data = ImageController.read(\"..\")\n```\nOutput:\n\n\n\nThe below sample code normalized the input image.\n```python\nfrom segmentationMVC.controller import ImageController\nnormalized_data = ImageController.normalize(image_data)\n```\nOutput:\n\n\n\n\nThe below sample code crops the input image.\n```python\nfrom segmentationMVC.controller import ImageController\ncrop_data = ImageController.normalize(normalized_data)\n```\nOutput:\n\n\n\n\nThe below sample code smoothens the input image.\n```python\nfrom segmentationMVC.controller import ImageController\nsmoothed_data = ImageController.smooth(crop_data)\n```\nOutput:\n\n\n\n\nTo get the complete segmented image, the following will serve as an example.\n```python\nfrom segmentationMVC.controller import ImageController\n\nimage_data = ImageController.read(\"..\")\n\nnormalized_data = ImageController.normalize(image_data)\n\ncrop_data = ImageController.normalize(normalized_data)\n\nsmoothed_data = ImageController.smooth(crop_data)\n\nthreshold_data = ImageController.threshold(smoothed_data, threshold_value=73)\nlabeled_data, nr_count = ImageController.label(threshold_data)\nselected_labeled_data = ImageController.select_regions(labeled_data, region_size=1500)\nbinary_mask_data = ImageController.binary_mask(selected_labeled_data)\nclosed_binary_mask_data = ImageController.close_binary_mask(binary_mask_data)\nsegmented_image = ImageController.binary_image(closed_binary_mask_data)\n```\nThe variable **segmented_image** in the above sample code stores the result of the segmented image.\n\n\nOutput:\n\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A package based on traditional segmentation algorithms used to segment a fly retina images.",
"version": "1.3.3",
"project_urls": {
"Bug Tracker": "https://github.com/kshvr16/CS410_FlyEM_PyPI/issues",
"Homepage": "https://github.com/kshvr16/CS410_FlyEM_PyPi"
},
"split_keywords": [
"medical image",
"medical image segmentation",
"fly retina images",
"electron microscopy images"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "816e308b677d1cc868649b8a298aeeb94559586c924f768e01c49c050b9da066",
"md5": "635d0a15aa0bb3344b667f4844030cfc",
"sha256": "e22a0f52862a21a10fd73cc24a1535ca04ae96e9bb7f086ca48d3b7b15c7e61e"
},
"downloads": -1,
"filename": "flyem_segmentation_pipeline-1.3.3.tar.gz",
"has_sig": false,
"md5_digest": "635d0a15aa0bb3344b667f4844030cfc",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 4774,
"upload_time": "2023-05-19T22:39:45",
"upload_time_iso_8601": "2023-05-19T22:39:45.422780Z",
"url": "https://files.pythonhosted.org/packages/81/6e/308b677d1cc868649b8a298aeeb94559586c924f768e01c49c050b9da066/flyem_segmentation_pipeline-1.3.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-05-19 22:39:45",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "kshvr16",
"github_project": "CS410_FlyEM_PyPi",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "flyem-segmentation-pipeline"
}
