# 近红外数据预处理流程
本项目包括整个近红外数据预处理流程,和相关算法。整体流程图如下所示:

## 一、光强数据读取和处理
所有以 raw 开头的函数文件名,均应用于光密度数据。
### 1.1 原始数据读取
```Python
# 读取原始光强数据
raw_df = raw_intensity_import(r"data/raw/raw_sample01.csv")
```
### 1.2 光强数据转换为光密度数据
提供以10为底和以自然对数为底两种方法,默认和推荐以10为底。
```Python
# 将原始光强数据转换为光密度数据 (默认 base10 对数)
od_df_base10 = raw_intensity_to_od(raw_df)
# 将原始光强数据转换为光密度数据 (以自然对数为底)
od_df_natural = raw_intensity_to_od(raw_df, base="natural")
```
### 1.3 光强数据的变异系数 CV
TODO
### 1.4 光强数据的信噪比 SNR
TODO
## 二、光密度数据读取和处理
### 2.1 光密度数据的读取
```Python
# 读取光密度数据并完成基线校正
od_df = OD_import(r"data/OD/OD_sample.csv")
```
### 2.2 光密度数据可视化
对光密度数据以通道进行子图可视化,示例图:
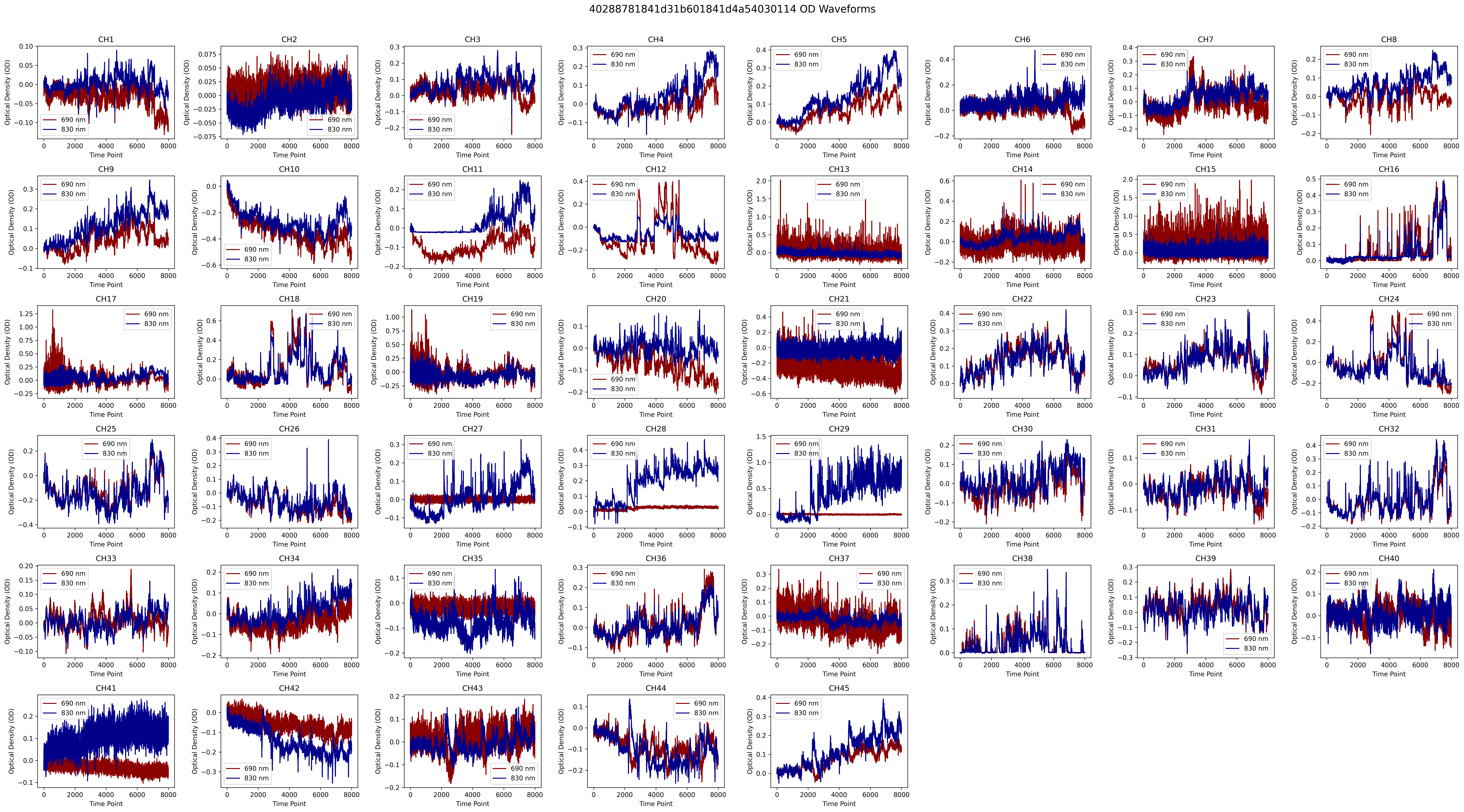
```Python
OD_visualize(od_df, "保存文件名", "output/figure")
```
### 2.3 计算各个通道头皮耦合指数(SCI)
可以自定义采样率和阈值。
```Python
sci_df = OD_scalp_coupling_index(od_df, sfreq=10, threshold=0.75)
```
### 2.4 光密度数据转换为血氧数据
```Python
HB_df_base10 = OD_beerlambert(od_df_base10, [690, 830], base='base10')
```
## 三、血氧数据处理
### 3.1 血氧数据的读取
```Python
HB_df = HB_import(r"data/HB/HB_sample02.csv")
```
### 3.2 血氧数据通道可视化
示例图:
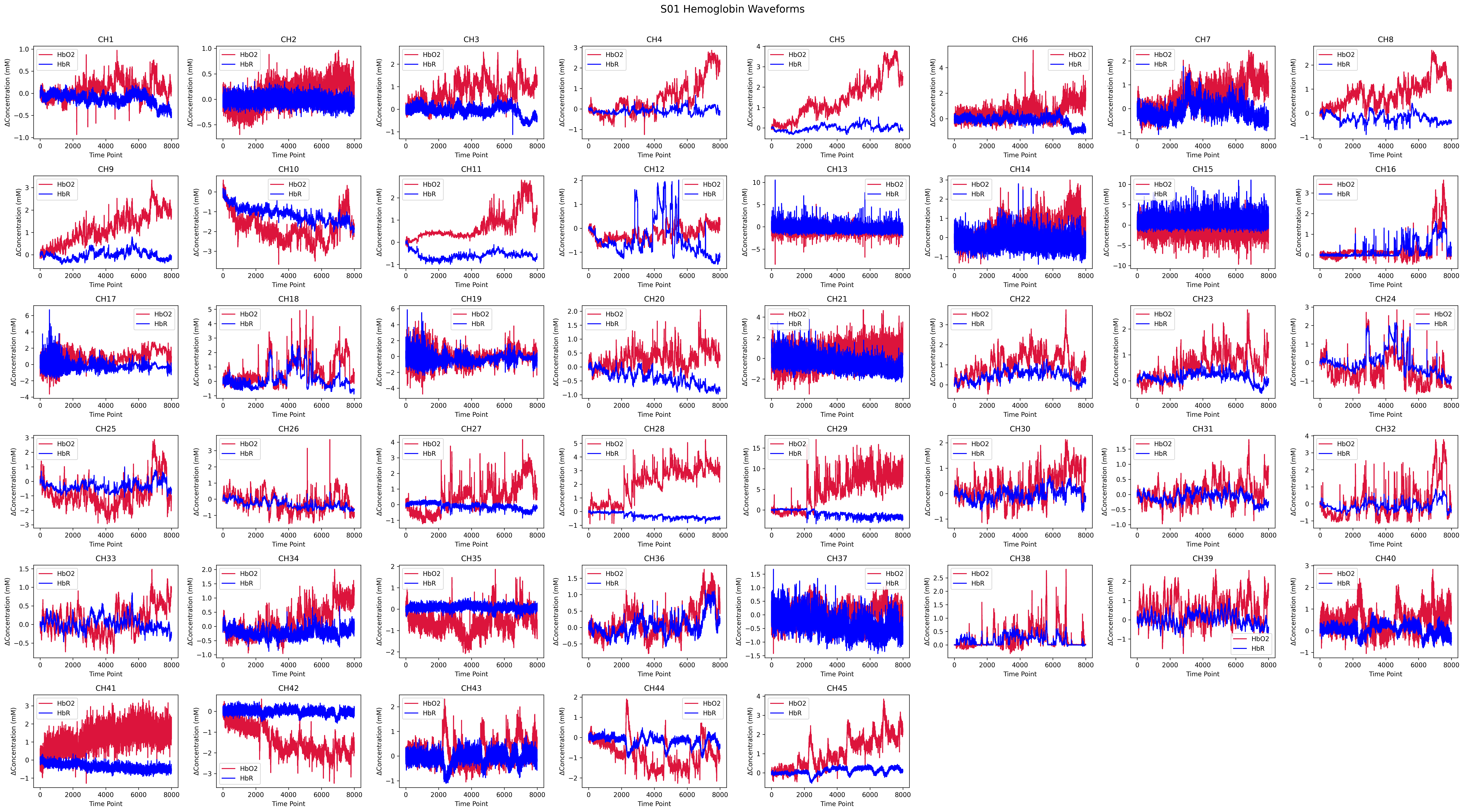
```Python
HB_visualize(data=hb_df, file_id=file_id, output_path="output/zlhk_figures/oxy",
title="S01 Hemoglobin Waveforms", mode='both', path_corrected=True)
```
### 3.3 血氧数据热图可视化
示例图:
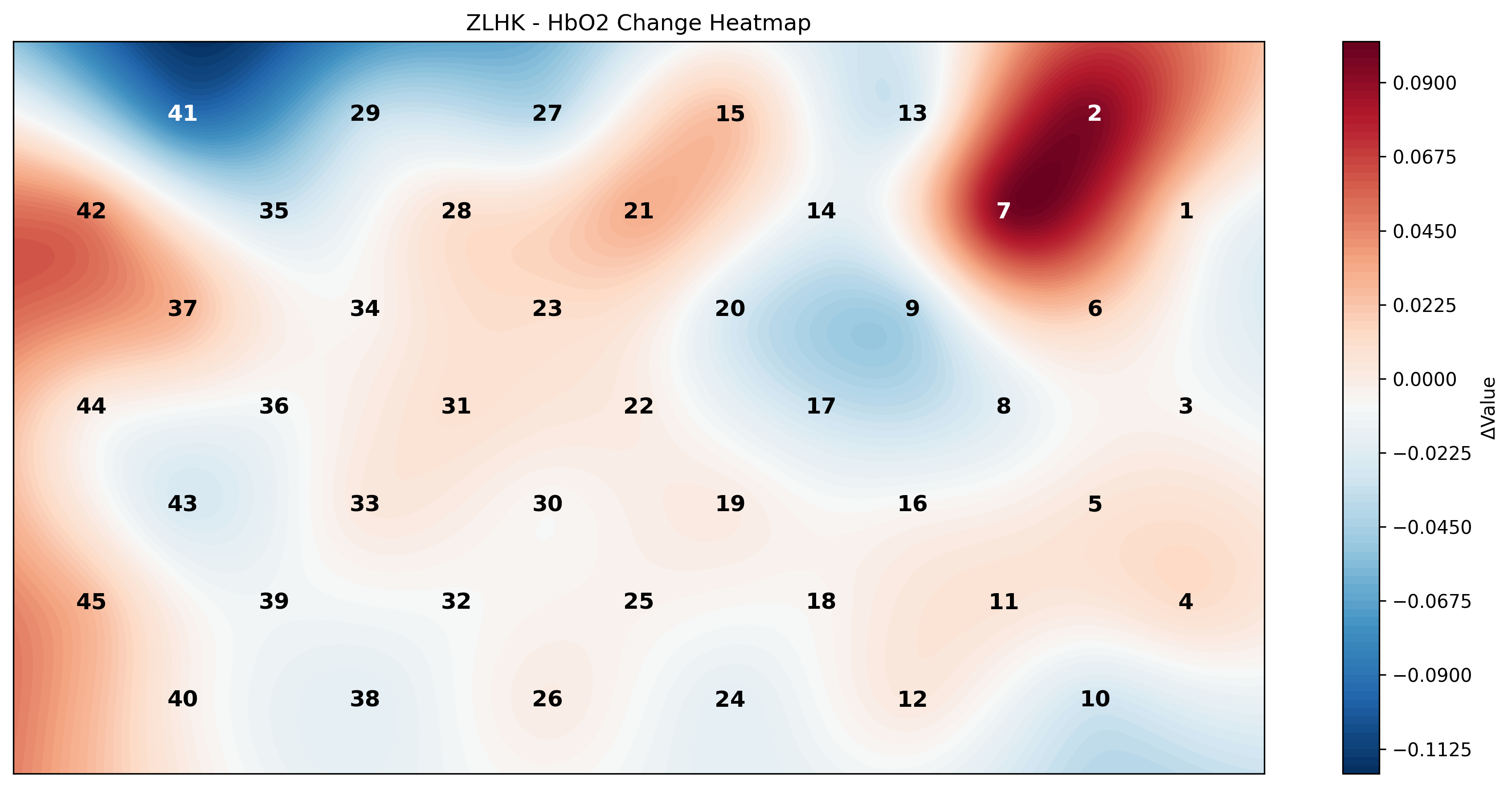
```Python
channel_df = pd.read_csv("./data/channel_location_ZLHK_Plate.csv")
HB_heatmap(channel_df, data_df,
signal_type="Oxy",
title="ZLHK - HbO2 Change Heatmap",
output_path = "output")
```
### 3.4 血氧数据通道 -> 脑区映射
```Python
region_map = json.load(open(r"data/channel_region_map.json"))
HB_brain_df = HB_brain_integration(HB_df, region_map, hb_type='oxy')
```
### 3.5 血氧脑区通道可视化
可视化示例图:
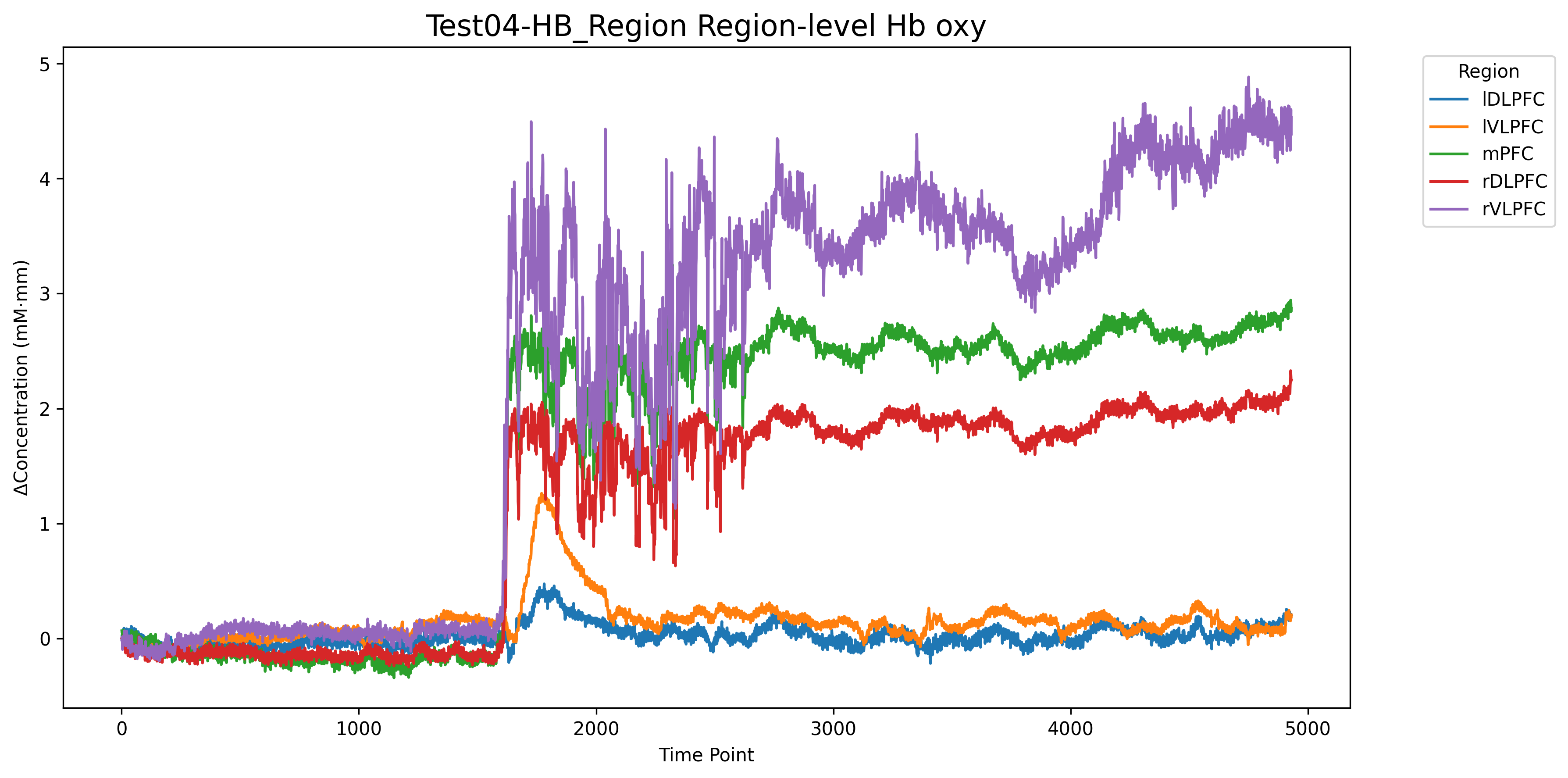
```Python
HB_brain_df = HB_brain_integration(HB_df, region_map, hb_type='oxy')
HB_region_visualize(HB_brain_df, file_name="Test04-HB_Region", output_path="output/figure",
title="", mode='oxy', path_corrected=False)
```
Raw data
{
"_id": null,
"home_page": "https://github.com/xzywisdili/fNIRSAnalysisProcess",
"name": "fnirs-toolkit",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": "Your Team <your.email@example.com>",
"keywords": "fnirs, neuroimaging, signal processing, hemodynamics, brain imaging",
"author": "Jamie, Wendy",
"author_email": "Your Team <your.email@example.com>",
"download_url": "https://files.pythonhosted.org/packages/71/e3/ae69d8c5751908f0a193afa4dae51b0a7cbafb02e4d9dfbde210dff7bba7/fnirs_toolkit-0.2.11.tar.gz",
"platform": null,
"description": "# \u8fd1\u7ea2\u5916\u6570\u636e\u9884\u5904\u7406\u6d41\u7a0b\r\n\r\n\u672c\u9879\u76ee\u5305\u62ec\u6574\u4e2a\u8fd1\u7ea2\u5916\u6570\u636e\u9884\u5904\u7406\u6d41\u7a0b\uff0c\u548c\u76f8\u5173\u7b97\u6cd5\u3002\u6574\u4f53\u6d41\u7a0b\u56fe\u5982\u4e0b\u6240\u793a\uff1a\r\n\r\n\r\n\r\n## \u4e00\u3001\u5149\u5f3a\u6570\u636e\u8bfb\u53d6\u548c\u5904\u7406\r\n\r\n\u6240\u6709\u4ee5 raw \u5f00\u5934\u7684\u51fd\u6570\u6587\u4ef6\u540d\uff0c\u5747\u5e94\u7528\u4e8e\u5149\u5bc6\u5ea6\u6570\u636e\u3002\r\n\r\n### 1.1 \u539f\u59cb\u6570\u636e\u8bfb\u53d6\r\n\r\n```Python\r\n# \u8bfb\u53d6\u539f\u59cb\u5149\u5f3a\u6570\u636e\r\nraw_df = raw_intensity_import(r\"data/raw/raw_sample01.csv\")\r\n```\r\n### 1.2 \u5149\u5f3a\u6570\u636e\u8f6c\u6362\u4e3a\u5149\u5bc6\u5ea6\u6570\u636e\r\n\r\n\u63d0\u4f9b\u4ee510\u4e3a\u5e95\u548c\u4ee5\u81ea\u7136\u5bf9\u6570\u4e3a\u5e95\u4e24\u79cd\u65b9\u6cd5\uff0c\u9ed8\u8ba4\u548c\u63a8\u8350\u4ee510\u4e3a\u5e95\u3002\r\n\r\n```Python\r\n# \u5c06\u539f\u59cb\u5149\u5f3a\u6570\u636e\u8f6c\u6362\u4e3a\u5149\u5bc6\u5ea6\u6570\u636e (\u9ed8\u8ba4 base10 \u5bf9\u6570)\r\nod_df_base10 = raw_intensity_to_od(raw_df)\r\n\r\n# \u5c06\u539f\u59cb\u5149\u5f3a\u6570\u636e\u8f6c\u6362\u4e3a\u5149\u5bc6\u5ea6\u6570\u636e (\u4ee5\u81ea\u7136\u5bf9\u6570\u4e3a\u5e95)\r\nod_df_natural = raw_intensity_to_od(raw_df, base=\"natural\")\r\n```\r\n### 1.3 \u5149\u5f3a\u6570\u636e\u7684\u53d8\u5f02\u7cfb\u6570 CV\r\n\r\nTODO\r\n\r\n### 1.4 \u5149\u5f3a\u6570\u636e\u7684\u4fe1\u566a\u6bd4 SNR\r\n\r\nTODO\r\n\r\n## \u4e8c\u3001\u5149\u5bc6\u5ea6\u6570\u636e\u8bfb\u53d6\u548c\u5904\u7406\r\n\r\n### 2.1 \u5149\u5bc6\u5ea6\u6570\u636e\u7684\u8bfb\u53d6\r\n\r\n```Python\r\n# \u8bfb\u53d6\u5149\u5bc6\u5ea6\u6570\u636e\u5e76\u5b8c\u6210\u57fa\u7ebf\u6821\u6b63\r\nod_df = OD_import(r\"data/OD/OD_sample.csv\")\r\n```\r\n\r\n### 2.2 \u5149\u5bc6\u5ea6\u6570\u636e\u53ef\u89c6\u5316\r\n\r\n\u5bf9\u5149\u5bc6\u5ea6\u6570\u636e\u4ee5\u901a\u9053\u8fdb\u884c\u5b50\u56fe\u53ef\u89c6\u5316\uff0c\u793a\u4f8b\u56fe\uff1a\r\n\r\n\r\n\r\n```Python\r\nOD_visualize(od_df, \"\u4fdd\u5b58\u6587\u4ef6\u540d\", \"output/figure\")\r\n```\r\n\r\n### 2.3 \u8ba1\u7b97\u5404\u4e2a\u901a\u9053\u5934\u76ae\u8026\u5408\u6307\u6570\uff08SCI\uff09\r\n\r\n\u53ef\u4ee5\u81ea\u5b9a\u4e49\u91c7\u6837\u7387\u548c\u9608\u503c\u3002\r\n\r\n```Python\r\nsci_df = OD_scalp_coupling_index(od_df, sfreq=10, threshold=0.75)\r\n```\r\n\r\n### 2.4 \u5149\u5bc6\u5ea6\u6570\u636e\u8f6c\u6362\u4e3a\u8840\u6c27\u6570\u636e\r\n\r\n```Python\r\nHB_df_base10 = OD_beerlambert(od_df_base10, [690, 830], base='base10')\r\n```\r\n\r\n## \u4e09\u3001\u8840\u6c27\u6570\u636e\u5904\u7406\r\n\r\n### 3.1 \u8840\u6c27\u6570\u636e\u7684\u8bfb\u53d6\r\n\r\n```Python\r\nHB_df = HB_import(r\"data/HB/HB_sample02.csv\")\r\n```\r\n\r\n### 3.2 \u8840\u6c27\u6570\u636e\u901a\u9053\u53ef\u89c6\u5316\r\n\r\n\u793a\u4f8b\u56fe\uff1a\r\n\r\n\r\n\r\n```Python\r\nHB_visualize(data=hb_df, file_id=file_id, output_path=\"output/zlhk_figures/oxy\",\r\n title=\"S01 Hemoglobin Waveforms\", mode='both', path_corrected=True)\r\n```\r\n\r\n### 3.3 \u8840\u6c27\u6570\u636e\u70ed\u56fe\u53ef\u89c6\u5316\r\n\r\n\u793a\u4f8b\u56fe\uff1a\r\n\r\n\r\n\r\n```Python\r\nchannel_df = pd.read_csv(\"./data/channel_location_ZLHK_Plate.csv\")\r\nHB_heatmap(channel_df, data_df,\r\n signal_type=\"Oxy\",\r\n title=\"ZLHK - HbO2 Change Heatmap\",\r\n output_path = \"output\")\r\n```\r\n\r\n### 3.4 \u8840\u6c27\u6570\u636e\u901a\u9053 -> \u8111\u533a\u6620\u5c04\r\n\r\n```Python\r\nregion_map = json.load(open(r\"data/channel_region_map.json\"))\r\nHB_brain_df = HB_brain_integration(HB_df, region_map, hb_type='oxy')\r\n```\r\n\r\n### 3.5 \u8840\u6c27\u8111\u533a\u901a\u9053\u53ef\u89c6\u5316\r\n\r\n\u53ef\u89c6\u5316\u793a\u4f8b\u56fe\uff1a\r\n\r\n\r\n\r\n```Python\r\nHB_brain_df = HB_brain_integration(HB_df, region_map, hb_type='oxy')\r\nHB_region_visualize(HB_brain_df, file_name=\"Test04-HB_Region\", output_path=\"output/figure\",\r\n title=\"\", mode='oxy', path_corrected=False)\r\n```\r\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A comprehensive Python package for fNIRS data analysis",
"version": "0.2.11",
"project_urls": {
"Bug Tracker": "https://github.com/yourusername/fnirs-toolkit/issues",
"Documentation": "https://fnirs-toolkit.readthedocs.io/",
"Homepage": "https://github.com/yourusername/fnirs-toolkit",
"Repository": "https://github.com/yourusername/fnirs-toolkit.git"
},
"split_keywords": [
"fnirs",
" neuroimaging",
" signal processing",
" hemodynamics",
" brain imaging"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "7a96ab061998e2e67454cd6a53761d4be0b03667d93756e86f5dc51680c6e1a9",
"md5": "493c5b3981c0c6acae01e3b3125146cd",
"sha256": "38bda1bd6c733bc7f9aa6eb453ac28ab3643a8a7814375c85c8702273b732d54"
},
"downloads": -1,
"filename": "fnirs_toolkit-0.2.11-py3-none-any.whl",
"has_sig": false,
"md5_digest": "493c5b3981c0c6acae01e3b3125146cd",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 58179,
"upload_time": "2025-08-21T10:25:11",
"upload_time_iso_8601": "2025-08-21T10:25:11.157160Z",
"url": "https://files.pythonhosted.org/packages/7a/96/ab061998e2e67454cd6a53761d4be0b03667d93756e86f5dc51680c6e1a9/fnirs_toolkit-0.2.11-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "71e3ae69d8c5751908f0a193afa4dae51b0a7cbafb02e4d9dfbde210dff7bba7",
"md5": "c6f015eb65d601040e4bb5f74dddbc9b",
"sha256": "cab7c8c5c68441950423cbf87eb3377c4f3dda105a5bb51d41de6c14f60301ab"
},
"downloads": -1,
"filename": "fnirs_toolkit-0.2.11.tar.gz",
"has_sig": false,
"md5_digest": "c6f015eb65d601040e4bb5f74dddbc9b",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 49331,
"upload_time": "2025-08-21T10:25:12",
"upload_time_iso_8601": "2025-08-21T10:25:12.928594Z",
"url": "https://files.pythonhosted.org/packages/71/e3/ae69d8c5751908f0a193afa4dae51b0a7cbafb02e4d9dfbde210dff7bba7/fnirs_toolkit-0.2.11.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-08-21 10:25:12",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "xzywisdili",
"github_project": "fNIRSAnalysisProcess",
"github_not_found": true,
"lcname": "fnirs-toolkit"
}
