Gecko is a Python library for the bulk generation and mutation of realistic personal data.
It is a spiritual successor to the GeCo framework which was initially published by Tran, Vatsalan and Christen.
Gecko reimplements the most promising aspects of the original framework for modern Python with a simplified API, adds
extra features and massively improves performance thanks to NumPy and Pandas.
# Installation
Install with pip:
```bash
pip install gecko-syndata
```
Install with [Poetry](https://python-poetry.org/):
```bash
poetry add gecko-syndata
```
# Basic usage
[Please see the docs for an in-depth guide on how to use the library.](https://ul-mds.github.io/gecko/)
Writing a data generation script with Gecko is usually split into two consecutive steps.
In the first step, data is generated based on information that you provide.
Most commonly, Gecko pulls the information it needs from frequency tables, although other means of generating data
are possible.
Gecko will then output a dataset to your specifications.
In the second step, a copy of this dataset is mutated.
Gecko provides functions which deliberately introduce errors into your dataset.
These errors can take shape in typos, edit errors and other common data sources.
By the end, you will have a generated dataset and a mutated copy thereof.
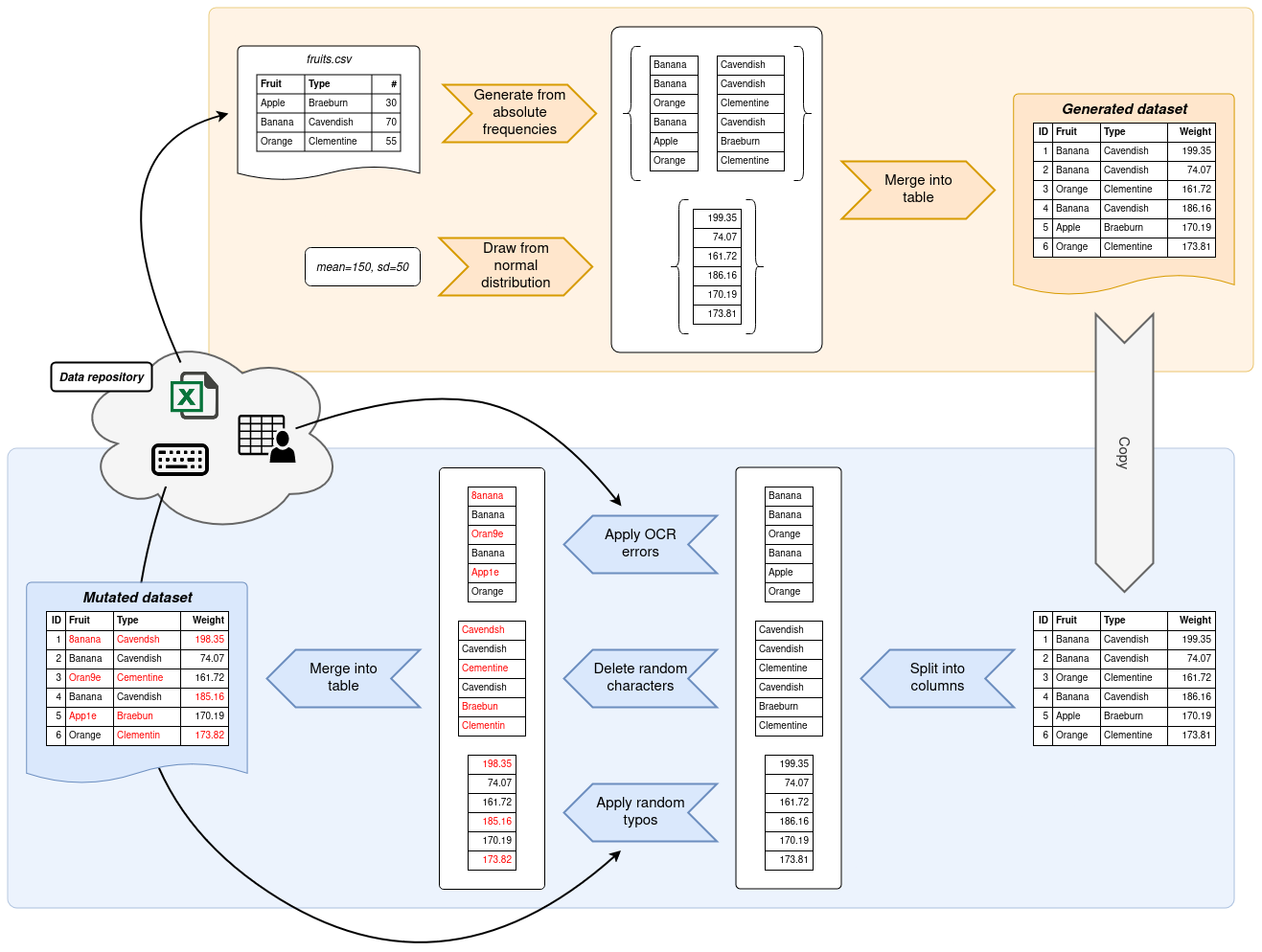
Gecko exposes two modules, `generator` and `mutator`, to help you write data generation scripts.
Both contain built-in functions covering the most common use cases for generating data from frequency information and
mutating data based on common error sources, such as typos, OCR errors and much more.
The following example gives a very brief overview of what a data generation script with Gecko might look like.
It uses frequency tables from the [Gecko data repository](https://github.com/ul-mds/gecko-data) which has been cloned
into a directory next to the script itself.
```python
from pathlib import Path
import numpy as np
from gecko import generator, mutator
# create a RNG with a set seed for reproducible results
rng = np.random.default_rng(727)
# path to the Gecko data repository
gecko_data_dir = Path("gecko-data")
# create a data frame with 10,000 rows and a single column called "last_name"
# which sources its values from the frequency table with the same name
df_generated = generator.to_data_frame(
[
("last_name", generator.from_frequency_table(
gecko_data_dir / "de_DE" / "last-name.csv",
value_column="last_name",
freq_column="count",
rng=rng,
)),
],
10_000,
)
# mutate this data frame by randomly deleting characters in 1% of all rows
df_mutated = mutator.mutate_data_frame(
df_generated,
[
("last_name", (.01, mutator.with_delete(rng))),
],
)
# export both data frames using Pandas' to_csv function
df_generated.to_csv("german-generated.csv", index_label="id")
df_mutated.to_csv("german-mutated.csv", index_label="id")
```
For a more extensive usage guide, [refer to the docs](https://ul-mds.github.io/gecko/).
# Rationale
The GeCo framework was originally conceived to facilitate the generation and mutation of personal data to validate
record linkage algorithms.
In the field of record linkage, acquiring real-world personal data to test new algorithms on is hard to come by.
Hence, GeCo went for a synthetic approach using statistical models from publicly available data.
GeCo was built for Python 2.7 and has not seen any active development since its last publication in 2013.
The general idea of providing shareable and reproducible Python scripts to generate personal data however still holds a
lot of promise.
This has led to the development of the Gecko library.
A lot of GeCo's weaknesses were rectified with this library.
Vectorized functions from Pandas and NumPy provide significant performance boosts and aid integration into existing
data science applications.
A simplified API allows for a much easier development of custom generators and mutators.
NumPy's random number generation routines instead of Python's built-in `random` module make fine-tuned reproducible
results a breeze.
Gecko therefore seeks to be GeCo's "bigger brother" and aims to provide a much more refined experience to generate
realistic personal data.
# Disclaimer
Gecko is still very much in a "beta" state.
As it stands, it satisfies our internal use cases within the Medical Data Science group, but we also seek wider
adoption.
If you find any issues or improvements with the library, do not hesitate to contact us.
# License
Gecko is released under the MIT License.
Raw data
{
"_id": null,
"home_page": "https://github.com/ul-mds/gecko",
"name": "gecko-syndata",
"maintainer": null,
"docs_url": null,
"requires_python": "<4.0,>=3.9",
"maintainer_email": null,
"keywords": "data science, data generation, data mutation",
"author": "Maximilian Jugl",
"author_email": "Maximilian.Jugl@medizin.uni-leipzig.de",
"download_url": "https://files.pythonhosted.org/packages/fe/0a/5a3fb7ec9bff923ec16741d4daff2f61225566efcaa6c20f729774e27264/gecko_syndata-0.6.3.tar.gz",
"platform": null,
"description": "Gecko is a Python library for the bulk generation and mutation of realistic personal data.\nIt is a spiritual successor to the GeCo framework which was initially published by Tran, Vatsalan and Christen.\nGecko reimplements the most promising aspects of the original framework for modern Python with a simplified API, adds\nextra features and massively improves performance thanks to NumPy and Pandas.\n\n# Installation\n\nInstall with pip:\n\n```bash\npip install gecko-syndata\n```\n\nInstall with [Poetry](https://python-poetry.org/):\n\n```bash\npoetry add gecko-syndata\n```\n\n# Basic usage\n\n[Please see the docs for an in-depth guide on how to use the library.](https://ul-mds.github.io/gecko/)\n\nWriting a data generation script with Gecko is usually split into two consecutive steps.\nIn the first step, data is generated based on information that you provide.\nMost commonly, Gecko pulls the information it needs from frequency tables, although other means of generating data\nare possible.\nGecko will then output a dataset to your specifications.\n\nIn the second step, a copy of this dataset is mutated.\nGecko provides functions which deliberately introduce errors into your dataset.\nThese errors can take shape in typos, edit errors and other common data sources.\nBy the end, you will have a generated dataset and a mutated copy thereof.\n\n\n\nGecko exposes two modules, `generator` and `mutator`, to help you write data generation scripts.\nBoth contain built-in functions covering the most common use cases for generating data from frequency information and\nmutating data based on common error sources, such as typos, OCR errors and much more.\n\nThe following example gives a very brief overview of what a data generation script with Gecko might look like.\nIt uses frequency tables from the [Gecko data repository](https://github.com/ul-mds/gecko-data) which has been cloned\ninto a directory next to the script itself.\n\n```python\nfrom pathlib import Path\n\nimport numpy as np\n\nfrom gecko import generator, mutator\n\n# create a RNG with a set seed for reproducible results\nrng = np.random.default_rng(727)\n# path to the Gecko data repository\ngecko_data_dir = Path(\"gecko-data\")\n\n# create a data frame with 10,000 rows and a single column called \"last_name\" \n# which sources its values from the frequency table with the same name\ndf_generated = generator.to_data_frame(\n [\n (\"last_name\", generator.from_frequency_table(\n gecko_data_dir / \"de_DE\" / \"last-name.csv\",\n value_column=\"last_name\",\n freq_column=\"count\",\n rng=rng,\n )),\n ],\n 10_000,\n)\n\n# mutate this data frame by randomly deleting characters in 1% of all rows\ndf_mutated = mutator.mutate_data_frame(\n df_generated,\n [\n (\"last_name\", (.01, mutator.with_delete(rng))),\n ],\n)\n\n# export both data frames using Pandas' to_csv function\ndf_generated.to_csv(\"german-generated.csv\", index_label=\"id\")\ndf_mutated.to_csv(\"german-mutated.csv\", index_label=\"id\")\n```\n\nFor a more extensive usage guide, [refer to the docs](https://ul-mds.github.io/gecko/).\n\n# Rationale\n\nThe GeCo framework was originally conceived to facilitate the generation and mutation of personal data to validate\nrecord linkage algorithms.\nIn the field of record linkage, acquiring real-world personal data to test new algorithms on is hard to come by.\nHence, GeCo went for a synthetic approach using statistical models from publicly available data.\nGeCo was built for Python 2.7 and has not seen any active development since its last publication in 2013.\nThe general idea of providing shareable and reproducible Python scripts to generate personal data however still holds a\nlot of promise.\nThis has led to the development of the Gecko library.\n\nA lot of GeCo's weaknesses were rectified with this library.\nVectorized functions from Pandas and NumPy provide significant performance boosts and aid integration into existing\ndata science applications.\nA simplified API allows for a much easier development of custom generators and mutators.\nNumPy's random number generation routines instead of Python's built-in `random` module make fine-tuned reproducible\nresults a breeze.\nGecko therefore seeks to be GeCo's \"bigger brother\" and aims to provide a much more refined experience to generate\nrealistic personal data.\n\n# Disclaimer\n\nGecko is still very much in a \"beta\" state.\nAs it stands, it satisfies our internal use cases within the Medical Data Science group, but we also seek wider\nadoption.\nIf you find any issues or improvements with the library, do not hesitate to contact us.\n\n# License\n\nGecko is released under the MIT License.\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Generation and mutation of realistic data at scale.",
"version": "0.6.3",
"project_urls": {
"Documentation": "https://ul-mds.github.io/gecko/",
"Homepage": "https://github.com/ul-mds/gecko",
"Repository": "https://github.com/ul-mds/gecko"
},
"split_keywords": [
"data science",
" data generation",
" data mutation"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "adb86fdaf203310136741e7f36d2b061dded760eac3000ce5bd1a4180d4a1211",
"md5": "744461ba7acf93f8df17cdcef50b7613",
"sha256": "87a00a82bef97d779daef952afbbb5dedc929d5567a7b8816df3d44fe1b999a6"
},
"downloads": -1,
"filename": "gecko_syndata-0.6.3-py3-none-any.whl",
"has_sig": false,
"md5_digest": "744461ba7acf93f8df17cdcef50b7613",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": "<4.0,>=3.9",
"size": 26254,
"upload_time": "2024-12-04T08:32:21",
"upload_time_iso_8601": "2024-12-04T08:32:21.316676Z",
"url": "https://files.pythonhosted.org/packages/ad/b8/6fdaf203310136741e7f36d2b061dded760eac3000ce5bd1a4180d4a1211/gecko_syndata-0.6.3-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "fe0a5a3fb7ec9bff923ec16741d4daff2f61225566efcaa6c20f729774e27264",
"md5": "0d46c6a776a75ce2cd89e362e00cab94",
"sha256": "6e498c92fba9bf13de0523d26e0a97ffa19d67b0560f9b4091976574dcd9f443"
},
"downloads": -1,
"filename": "gecko_syndata-0.6.3.tar.gz",
"has_sig": false,
"md5_digest": "0d46c6a776a75ce2cd89e362e00cab94",
"packagetype": "sdist",
"python_version": "source",
"requires_python": "<4.0,>=3.9",
"size": 25687,
"upload_time": "2024-12-04T08:32:22",
"upload_time_iso_8601": "2024-12-04T08:32:22.301234Z",
"url": "https://files.pythonhosted.org/packages/fe/0a/5a3fb7ec9bff923ec16741d4daff2f61225566efcaa6c20f729774e27264/gecko_syndata-0.6.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-12-04 08:32:22",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "ul-mds",
"github_project": "gecko",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "gecko-syndata"
}
