# Welcome
Contact : david.simonne@universite-paris-saclay.fr
You can install the latest version of the package by cloning the repository and via the `setup.py` script (`pip install .`)
* You can install gwaihir by typing `pip install -U gwaihir` in your command line. A stable version from the master branch uploaded to pypi.org will be used (`https://pypi.org/project/gwaihir/`)
* Otherwise, you can install the latest commit of the package by cloning this repository and typing `pip install .` in the terminal (you must be in the same directory as the `setup.py` script), this will allow you to have the latest updates.
Here is a link to a poster that tries to present Gwaihir:
[Poster_Gwaihir.pdf](https://www.dsimonne.eu/PhDAttachments/Poster_Gwaihir.pdf)
And to the paper: [Simonne, D., Carnis, J., Atlan, C., Chatelier, C., Favre-Nicolin, V., Dupraz, M., Leake, S. J., Zatterin, E., Resta, A., Coati, A. & Richard, M. I. (2022). J. Appl. Cryst. 55, 1045-1054](https://scripts.iucr.org/cgi-bin/paper?S1600576722005854)
## Important code snippets
To increase the width of the cells in Jupyter Notebook:
```python
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:75% !important; }</style>"))
```
To avoid automatic cell scrolling:
```javascript
%%javascript
IPython.OutputArea.prototype._should_scroll = function(lines) {
return false;
}
```
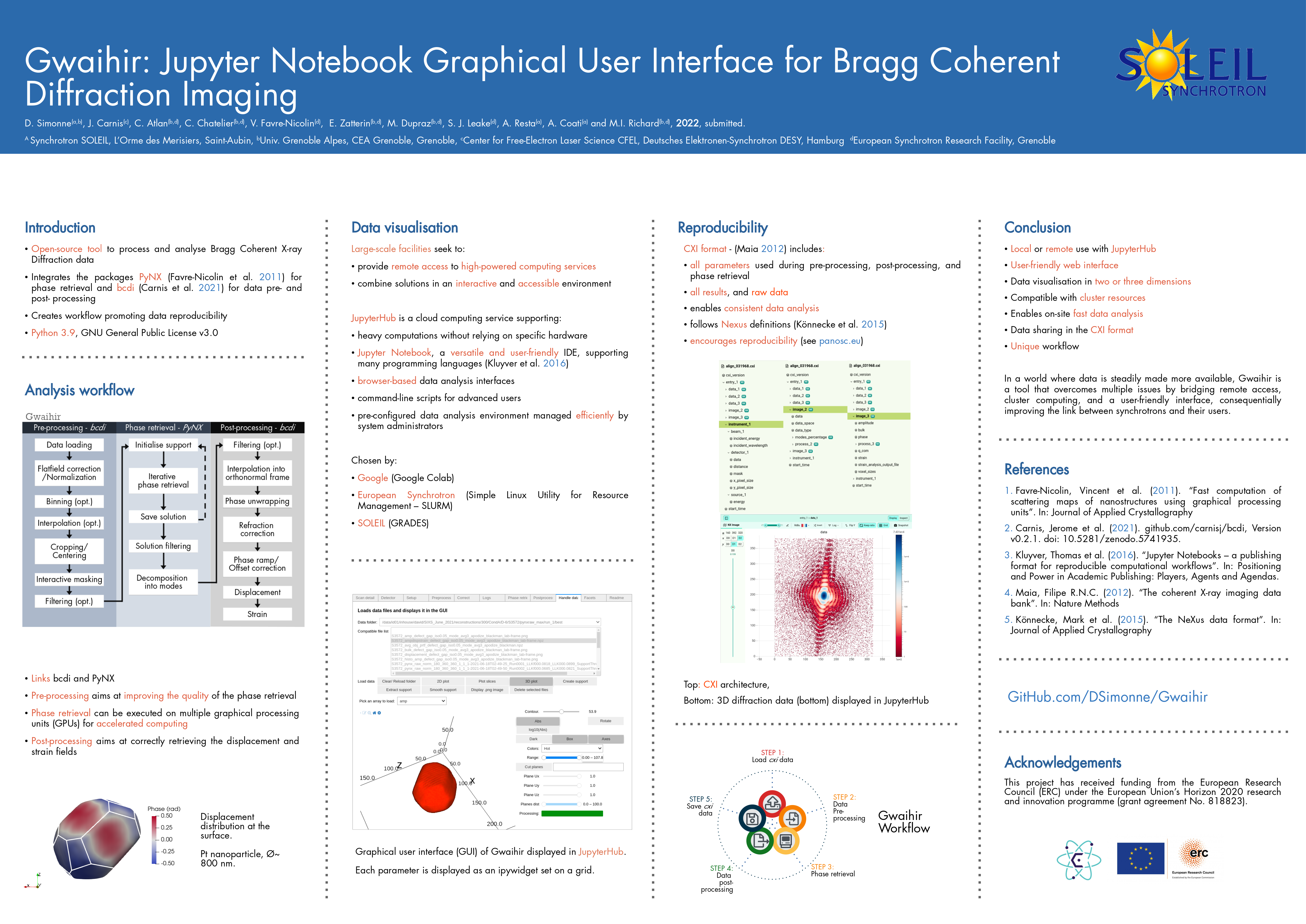
To open the GUI:
```python
from gwaihir.gui import Interface
GUI = Interface()
```
# GUI Preview:
## Pre-processing data
https://user-images.githubusercontent.com/51970962/154160601-f3e7878a-d2c6-4560-95e5-adf7087f59ab.mp4
## Phase retrieval
https://user-images.githubusercontent.com/51970962/154160830-f3c6460b-14e5-4bcc-99f5-e8691278a4e9.mp4
## Data plotting
https://user-images.githubusercontent.com/51970962/154160549-c5caea1b-afa0-4a29-a5a8-aff8a1a5158b.mp4
## Post-processing
https://user-images.githubusercontent.com/51970962/154236802-24643473-1ee9-4d01-823c-beca07ea1c58.mp4
An example file can be downloaded at: https://www.dsimonne.eu/Attachments/align_031968.cxi
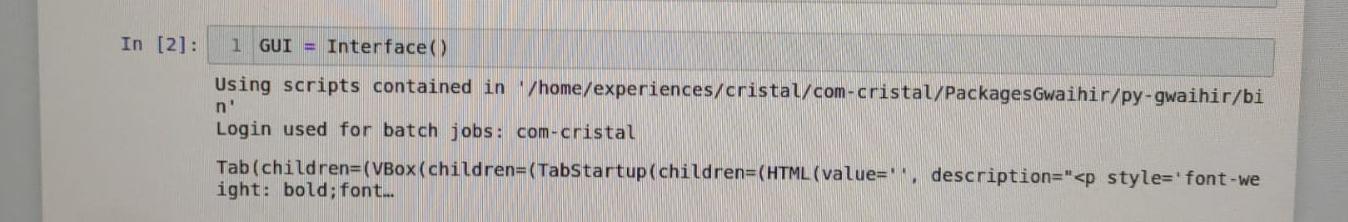
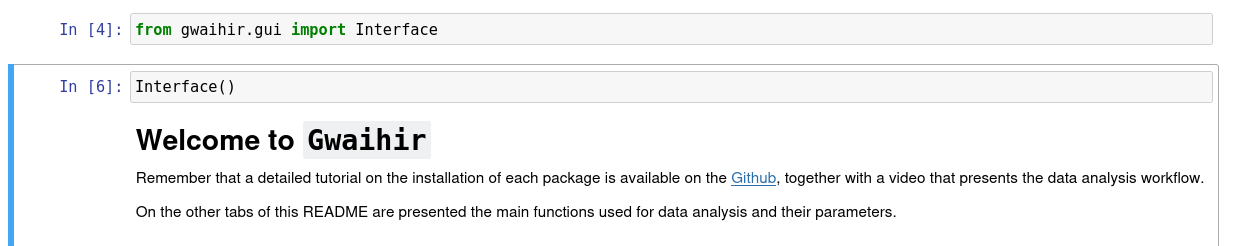
# Known bog
The first time you install `gwaihir`, it is possible that when you open the Interface, you only see some text printed, or the content of the readme tab, but you cannot interact with anything. This is solved by restarting your computer.
Bog with printed text:
Bog with README tab:
# Clusters at ESRF
Gwaihir **only** works with the p9 partition at the ESRF, optimized for phase retrieval.
If you want to use it for data analysis, you can install `gwaihir` and `bcdi` on rnice.
## Jupyter-slurm
How to access:
* Web browser: https://jupyter-slurm.esrf.fr/hub/spawn
* Terminal (for advanced users) :
* `ssh -X <login>@slurm-nice-devel`
* Ask for a GPU: `srun -N 1 --partition=p9gpu --gres=gpu:1 --time=06:00:00 --pty bash`
#### Available environements
* `/usr/bin/python3`: your personal environemnt
* p9.dev : optimised for BCDI, gwaihir and PyNX, development version, `source /data/id01/inhouse/david/p9.dev/bin/activate`
* p9.stable : optimised for BCDI, gwaihir and PyNX, stable version, `source /data/id01/inhouse/david/p9.stable/bin/activate`
* p9.pynx-devel : pynx only, frequently updated : `source /sware/exp/pynx/devel.p9/bin/activate`
You are not allowed to **modify** these environments but you can **link** a kernel if you wish to **use** them in jupyter.
To do so:
* Source the environment; e.g. `source /data/id01/inhouse/david/p9.dev/bin/activate`
* Make sure that:
* you are on slurm
* you requested a GPU
* Create the kernel:
* `python3 -m ipykernel install --user --name p9.stable`
* [Documentation](https://ipython.readthedocs.io/en/stable/install/kernel_install.html)
Once you feel confident, you should create your own environment, to avoid sudden updates that may impact your work!
To list the kernels you have installed: `jupyter kernelspec list`
And to remove them: `jupyter kernelspec uninstall <kernelname>`
Make sure that you are using the right kernel on your Jupyter Notebook !
### Set up `ssh` connection without using password (mandatory for batch jobs)
* Login into slurm (make sure that you asked for a GPU)
* Open a terminal (new -> terminal)
Enter the following commands (replace `<username>` with your username, for me it is simonne)
* `cd`
* `ssh-keygen -t rsa` (press enter when prompted, ~ 3 times)
* `ssh <username>@slurm-nice-devel mkdir -p .ssh`
* `cat .ssh/id_rsa.pub | ssh <username>@slurm-nice-devel 'cat >> .ssh/authorized_keys'`
You should not need a password anymore when login into slurm, make sure it is the case by typing
* `ssh <username>@slurm-nice-devel`
# CLuster at SOLEIL
## GRADES
To analyse data recorded at SOLEIL from your personal computer, you can use Jupyter Notebook via GRADES. The documentation is here (accessible on- site via the SOLEIL wifi or with the SOLEIL VPN) : http://confluence.synchrotron-soleil.fr/display/EG/Service%3A+Jupyter+Notebook
* Use this link to open Jupyter Notebook : http://grades-01.synchrotron-soleil.fr/notebook/
* Launch a Terminal (command line) from the upper-right 'New' menu. The system is a Debian 10, so you can enter linux commands there.
`PyNX` is already installed on GRADES, a version that is out of my control. So you 'just' have to download the `bcdi` and `gwaihir` packages by typing `pip3 install --proxy=http://195.221.0.35:8080 -U gwaihir bcdi`
If you encounter an error with gwaihir or bcdi, it is possible that the pip packages are not up to date. Then you should follow the procedure described [below](https://github.com/DSimonne/gwaihir#installing-different-packages-yourself) and manually download the packages. Just replace `pip install` by `pip3 install --proxy=http://195.221.0.35:8080`, the proxy IP can be 195.221.0.34:8080 or 195.221.0.35:8080 on the ReS (offices and VPN), and 195.221.10.6:8080 or 195.221.10.7:8080 on the REL (beam-lines, RAS).
You can also directly use a virtual machine provided by GRADES
* Go to https://re-grades-01.exp.synchrotron-soleil.fr/qemu-web-desktop/
* Use the SUNset/LDAP id.
* Click on `Create` then on `Connect` (after 10 sec).
* Once the desktop is available, search "gwaihir" in the bottom left menu and execute the program.
* You are now connected to the ruche with an environment that has all the necessary packages, you just need to open a notebook.
## SixS
A GPU is installed on sixs3, a computer available on the beamline, for phase retrieval.
Please respect the following steps:
* Make sure that you are logged in as `com-sixs`
* Activate the environment `source_py3.9` or `source /home/experiences/sixs/simonne/Documents/py39-env/bin/activate`, this environment is protected and you cannot modify it.
* Launch `jupyter notebook`
* Go to the test_data folder and then choose the beamline you want to test
* Follow the instructions in the notebook
## Cristal
A GPU is installed on cristal4, a computer available on the beamline, for phase retrieval.
Please respect the following steps:
* Make sure that you are logged in as `com-cristal`
* Activate the environment `source_gwaihir` or `source /home/experiences/crystal/com-cristal/PackagesGwaihir/py-gwaihir/bin/activate`, this environment is protected and you cannot modify it.
* Launch `jupyter notebook`
* Go to the test_data folder and then choose the beamline you want to test
* Follow the instructions in the notebook
# Installing different packages yourself
* First, I advise you to create a `/Packages` directory to keep these.
* Secondly, I advise you to create a virtual environment to help with debogging, and so that once everything works, you don't update a package by mistake. To do so please follow the following steps:
## Create a virtual environment
* `mkdir py38-env`
* `cd py38-env/`
* `python3.8 -m venv .`
* `source bin/activate` # To activate the environment
* Make sure `wheel` and `setuptools` are installed: `pip install wheel setuptools pip --upgrade`
Then you should create an alias such as: `alias source_p9="source /home/user/py38-env/bin/activate"`
## Specific instructions for the p.9 cluster
* If `vtk` does not install (on the p9 cluster at the ESRF for example), you can type : `pip install --trusted-host www.silx.org --find-links http://www.silx.org/pub/wheelhouse vtk`, you may also need to remove the version requirements in `bcdi/setup.py`
* If `PyQt5` does not install (also on the p9 cluster at the ESRF), you can install it by activating your environment from the rnice cluster.
## 1) Install PyNX
* Use the latest version
* `cd /Packages`
* `mkdir PyNX_install`
* `cd PyNX_install/`
* `curl -O http://ftp.esrf.fr/pub/scisoft/PyNX/pynx-devel-nightly.tar.bz2` # Installation details within install-pynx-venv.sh
* `source_p9`
* `pip install pynx-devel-nightly.tar.bz2[cuda,gui,mpi]` # Install with extras cuda, mpi, cdi
* cite `PyNX: high-performance computing toolkit for coherent X-ray imaging based on operators is out: J. Appl. Cryst. 53 (2020), 1404`, also available as `arXiv:2008.11511`
## 2) Install gwaihir
* `cd /Packages`
* `git clone https://github.com/DSimonne/gwaihir.git`
* `cd gwaihir`
* `source_p9`
* `pip install .`
* cite `Simonne, D., Carnis, J., Atlan, C., Chatelier, C., Favre-Nicolin, V., Dupraz, M., Leake, S. J., Zatterin, E., Resta, A., Coati, A. & Richard, M. I. (2022). J. Appl. Cryst. 55, 1045-1054.`
## 3) Install bcdi
* Latest version tested : v0.2.8
* `cd /Packages`
* `git clone https://github.com/carnisj/bcdi.git`
* `cd bcdi`
* `source_p9`
* `pip install .`
* cite `DOI: 10.5281/zenodo.3257616`
## 4) Install facet-analyser (Debian 11 only)
* Send a thank you email to Fred Picca =D
* `cd /Packages`
* `git clone https://salsa.debian.org/science-team/facet-analyser.git`
* `cd facet-analyser`
* `git checkout`
* `sudo mk-build-deps -i`
* Make sure that you have qt installed, for me I had to install `libqt5opengl5-dev` (debian-testing)
* `debuild -b`
* if the package creation fail, try to ignore the test in /debian/rules (line 19)
* `sudo debi`
* The package is now installed. You can check the locations of its files with the command `dpkg -L facet-analyser`
* You should see a file named `/usr/lib/x86_64-linux-gnu/paraview-5.9/plugins/FacetAnalyser/FacetAnalyser.so`
* Now launch `/usr/bin/paraview` (if not installed yet, good luck, refer to `https://www.paraview.org/Wiki/ParaView:Build_And_Install#Installing`)
* In paraview, go to Tools > Manage Plugins > Load New
* Here type the path to the plugin that was printed with the `dpkg -L facet-analyser` command.
* Feel free to add it to `/usr/bin/plugin` so that it is loaded automatically.
* cite `Grothausmann, R. (2015). Facet Analyser : ParaView plugin for automated facet detection and measurement of interplanar angles of tomographic objects. March.`
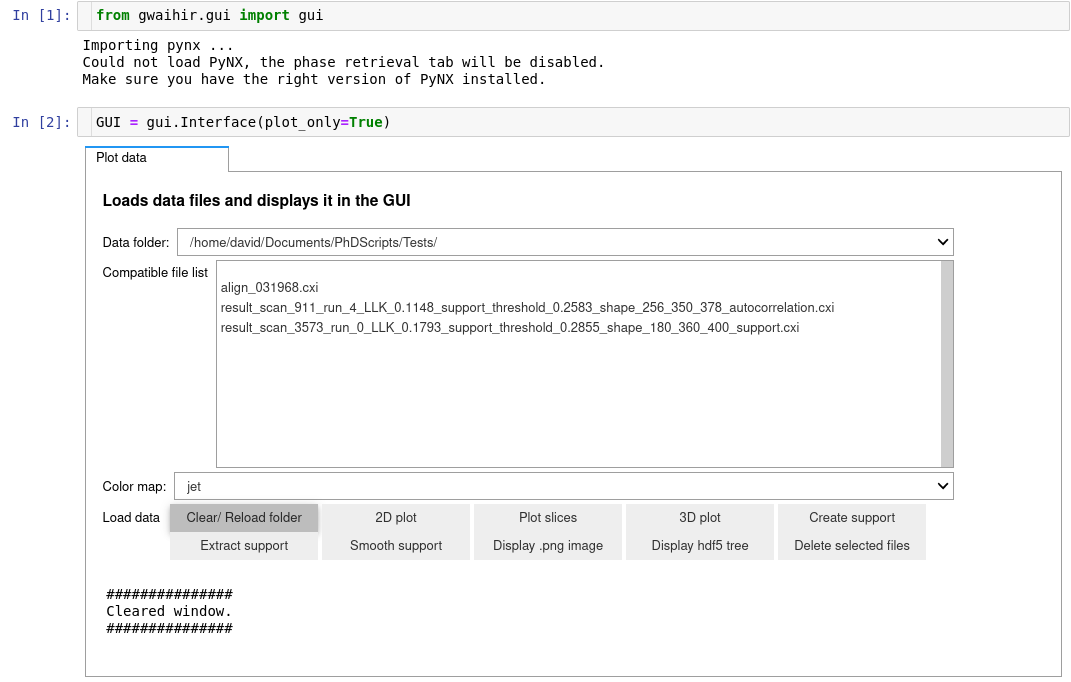
# To go further ...
## Using `Gwaihir` only as a plotting tool in Jupyter Notebook
## Quick navigation between `vtk` files in the GUI (outdated but it can give you some ideas)
It is possible to automate the navigation in the GUI !
Here I have a pandas DataFrame that contains data about my scans, I use to automate the navigation:
```python
import time
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import glob
df = pd.read_csv("reconstructions/scans_data.csv")
GUI.tab_facet.children[3].value = False
GUI.window.selected_index = 0
time.sleep(1)
GUI._list_widgets_init_dir.children[7].value = False
time.sleep(1)
scan = 3600
row = df[df.scan == scan]
particle = row.particle.values[0]
temp = row.temp_given.values[0]
condition = row.condition.values[0]
GUI._list_widgets_init_dir.children[2].value = scan
GUI._list_widgets_init_dir.children[
3].value = f"/data/id01/inhouse/david/SIXS_June_2021/reconstructions/{temp}/{condition}/{particle}/S{scan}/data/"
GUI._list_widgets_init_dir.children[
4].value = f"/data/id01/inhouse/david/SIXS_June_2021/reconstructions/{temp}/{condition}/{particle}/"
time.sleep(1)
GUI._list_widgets_init_dir.children[7].value = True
time.sleep(1)
GUI.window.selected_index = 9
GUI.tab_facet.children[3].value = "load_csv"
```
Raw data
{
"_id": null,
"home_page": "https://github.com/DSimonne/gwaihir/tree/master",
"name": "gwaihir",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.6",
"maintainer_email": "",
"keywords": "BCDI,ipywidgets,PyNX",
"author": "David Simonne",
"author_email": "david.simonne@universite-paris-saclay.fr",
"download_url": "https://files.pythonhosted.org/packages/c5/8f/b6c42fd4975d655b0c2c384abddfe69040449c24495594658b43c80758a4/gwaihir-0.0.6.tar.gz",
"platform": null,
"description": "# Welcome \n\nContact : david.simonne@universite-paris-saclay.fr\n\nYou can install the latest version of the package by cloning the repository and via the `setup.py` script (`pip install .`)\n\n* You can install gwaihir by typing `pip install -U gwaihir` in your command line. A stable version from the master branch uploaded to pypi.org will be used (`https://pypi.org/project/gwaihir/`)\n* Otherwise, you can install the latest commit of the package by cloning this repository and typing `pip install .` in the terminal (you must be in the same directory as the `setup.py` script), this will allow you to have the latest updates.\n\nHere is a link to a poster that tries to present Gwaihir:\n[Poster_Gwaihir.pdf](https://www.dsimonne.eu/PhDAttachments/Poster_Gwaihir.pdf)\n\nAnd to the paper: [Simonne, D., Carnis, J., Atlan, C., Chatelier, C., Favre-Nicolin, V., Dupraz, M., Leake, S. J., Zatterin, E., Resta, A., Coati, A. & Richard, M. I. (2022). J. Appl. Cryst. 55, 1045-1054](https://scripts.iucr.org/cgi-bin/paper?S1600576722005854)\n\n\n\n\n## Important code snippets\n\nTo increase the width of the cells in Jupyter Notebook:\n\n```python\nfrom IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container { width:75% !important; }</style>\"))\n```\n\nTo avoid automatic cell scrolling:\n\n```javascript\n%%javascript\nIPython.OutputArea.prototype._should_scroll = function(lines) {\n return false;\n}\n```\n\nTo open the GUI:\n\n```python\nfrom gwaihir.gui import Interface\nGUI = Interface()\n```\n\n\n# GUI Preview:\n## Pre-processing data\nhttps://user-images.githubusercontent.com/51970962/154160601-f3e7878a-d2c6-4560-95e5-adf7087f59ab.mp4\n\n## Phase retrieval\nhttps://user-images.githubusercontent.com/51970962/154160830-f3c6460b-14e5-4bcc-99f5-e8691278a4e9.mp4\n\n## Data plotting\nhttps://user-images.githubusercontent.com/51970962/154160549-c5caea1b-afa0-4a29-a5a8-aff8a1a5158b.mp4\n\n## Post-processing\nhttps://user-images.githubusercontent.com/51970962/154236802-24643473-1ee9-4d01-823c-beca07ea1c58.mp4\n\nAn example file can be downloaded at: https://www.dsimonne.eu/Attachments/align_031968.cxi\n\n# Known bog\n\nThe first time you install `gwaihir`, it is possible that when you open the Interface, you only see some text printed, or the content of the readme tab, but you cannot interact with anything. This is solved by restarting your computer.\n\nBog with printed text:\n\n\nBog with README tab:\n\n\n# Clusters at ESRF\n\nGwaihir **only** works with the p9 partition at the ESRF, optimized for phase retrieval.\n\nIf you want to use it for data analysis, you can install `gwaihir` and `bcdi` on rnice.\n\n## Jupyter-slurm\n\nHow to access:\n* Web browser: https://jupyter-slurm.esrf.fr/hub/spawn\n* Terminal (for advanced users) :\n * `ssh -X <login>@slurm-nice-devel`\n * Ask for a GPU: `srun -N 1 --partition=p9gpu --gres=gpu:1 --time=06:00:00 --pty bash`\n\n#### Available environements\n* `/usr/bin/python3`: your personal environemnt\n* p9.dev : optimised for BCDI, gwaihir and PyNX, development version, `source /data/id01/inhouse/david/p9.dev/bin/activate`\n* p9.stable : optimised for BCDI, gwaihir and PyNX, stable version, `source /data/id01/inhouse/david/p9.stable/bin/activate`\n* p9.pynx-devel : pynx only, frequently updated : `source /sware/exp/pynx/devel.p9/bin/activate`\n\nYou are not allowed to **modify** these environments but you can **link** a kernel if you wish to **use** them in jupyter.\n\nTo do so:\n* Source the environment; e.g. `source /data/id01/inhouse/david/p9.dev/bin/activate`\n* Make sure that:\n * you are on slurm\n * you requested a GPU\n* Create the kernel:\n * `python3 -m ipykernel install --user --name p9.stable`\n* [Documentation](https://ipython.readthedocs.io/en/stable/install/kernel_install.html)\n\nOnce you feel confident, you should create your own environment, to avoid sudden updates that may impact your work!\n\nTo list the kernels you have installed: `jupyter kernelspec list`\n\nAnd to remove them: `jupyter kernelspec uninstall <kernelname>`\n\nMake sure that you are using the right kernel on your Jupyter Notebook !\n\n### Set up `ssh` connection without using password (mandatory for batch jobs)\n* Login into slurm (make sure that you asked for a GPU)\n* Open a terminal (new -> terminal)\n\nEnter the following commands (replace `<username>` with your username, for me it is simonne)\n* `cd`\n* `ssh-keygen -t rsa` (press enter when prompted, ~ 3 times)\n* `ssh <username>@slurm-nice-devel mkdir -p .ssh`\n* `cat .ssh/id_rsa.pub | ssh <username>@slurm-nice-devel 'cat >> .ssh/authorized_keys'`\n\nYou should not need a password anymore when login into slurm, make sure it is the case by typing\n* `ssh <username>@slurm-nice-devel`\n\n# CLuster at SOLEIL\n\n## GRADES\n\nTo analyse data recorded at SOLEIL from your personal computer, you can use Jupyter Notebook via GRADES. The documentation is here (accessible on- site via the SOLEIL wifi or with the SOLEIL VPN) : http://confluence.synchrotron-soleil.fr/display/EG/Service%3A+Jupyter+Notebook\n\n* Use this link to open Jupyter Notebook : http://grades-01.synchrotron-soleil.fr/notebook/\n* Launch a Terminal (command line) from the upper-right 'New' menu. The system is a Debian 10, so you can enter linux commands there.\n\n`PyNX` is already installed on GRADES, a version that is out of my control. So you 'just' have to download the `bcdi` and `gwaihir` packages by typing `pip3 install --proxy=http://195.221.0.35:8080 -U gwaihir bcdi`\n\nIf you encounter an error with gwaihir or bcdi, it is possible that the pip packages are not up to date. Then you should follow the procedure described [below](https://github.com/DSimonne/gwaihir#installing-different-packages-yourself) and manually download the packages. Just replace `pip install` by `pip3 install --proxy=http://195.221.0.35:8080`, the proxy IP can be 195.221.0.34:8080 or 195.221.0.35:8080 on the ReS (offices and VPN), and 195.221.10.6:8080 or 195.221.10.7:8080 on the REL (beam-lines, RAS).\n\nYou can also directly use a virtual machine provided by GRADES\n* Go to https://re-grades-01.exp.synchrotron-soleil.fr/qemu-web-desktop/\n* Use the SUNset/LDAP id.\n* Click on `Create` then on `Connect` (after 10 sec).\n* Once the desktop is available, search \"gwaihir\" in the bottom left menu and execute the program.\n* You are now connected to the ruche with an environment that has all the necessary packages, you just need to open a notebook.\n\n## SixS\n\nA GPU is installed on sixs3, a computer available on the beamline, for phase retrieval.\n\nPlease respect the following steps:\n* Make sure that you are logged in as `com-sixs`\n* Activate the environment `source_py3.9` or `source /home/experiences/sixs/simonne/Documents/py39-env/bin/activate`, this environment is protected and you cannot modify it.\n* Launch `jupyter notebook`\n* Go to the test_data folder and then choose the beamline you want to test\n* Follow the instructions in the notebook\n\n## Cristal\n\nA GPU is installed on cristal4, a computer available on the beamline, for phase retrieval.\n\nPlease respect the following steps:\n* Make sure that you are logged in as `com-cristal`\n* Activate the environment `source_gwaihir` or `source /home/experiences/crystal/com-cristal/PackagesGwaihir/py-gwaihir/bin/activate`, this environment is protected and you cannot modify it.\n* Launch `jupyter notebook`\n* Go to the test_data folder and then choose the beamline you want to test\n* Follow the instructions in the notebook\n\n# Installing different packages yourself\n\n* First, I advise you to create a `/Packages` directory to keep these.\n* Secondly, I advise you to create a virtual environment to help with debogging, and so that once everything works, you don't update a package by mistake. To do so please follow the following steps:\n\n## Create a virtual environment\n\n* `mkdir py38-env`\n* `cd py38-env/`\n* `python3.8 -m venv .`\n* `source bin/activate` # To activate the environment\n* Make sure `wheel` and `setuptools` are installed: `pip install wheel setuptools pip --upgrade`\n\nThen you should create an alias such as: `alias source_p9=\"source /home/user/py38-env/bin/activate\"`\n\n## Specific instructions for the p.9 cluster\n* If `vtk` does not install (on the p9 cluster at the ESRF for example), you can type : `pip install --trusted-host www.silx.org --find-links http://www.silx.org/pub/wheelhouse vtk`, you may also need to remove the version requirements in `bcdi/setup.py`\n* If `PyQt5` does not install (also on the p9 cluster at the ESRF), you can install it by activating your environment from the rnice cluster.\n\n## 1) Install PyNX\n* Use the latest version\n* `cd /Packages`\n* `mkdir PyNX_install`\n* `cd PyNX_install/`\n* `curl -O http://ftp.esrf.fr/pub/scisoft/PyNX/pynx-devel-nightly.tar.bz2` # Installation details within install-pynx-venv.sh\n* `source_p9`\n* `pip install pynx-devel-nightly.tar.bz2[cuda,gui,mpi]` # Install with extras cuda, mpi, cdi\n* cite `PyNX: high-performance computing toolkit for coherent X-ray imaging based on operators is out: J. Appl. Cryst. 53 (2020), 1404`, also available as `arXiv:2008.11511`\n\n## 2) Install gwaihir\n* `cd /Packages`\n* `git clone https://github.com/DSimonne/gwaihir.git`\n* `cd gwaihir`\n* `source_p9`\n* `pip install .`\n* cite `Simonne, D., Carnis, J., Atlan, C., Chatelier, C., Favre-Nicolin, V., Dupraz, M., Leake, S. J., Zatterin, E., Resta, A., Coati, A. & Richard, M. I. (2022). J. Appl. Cryst. 55, 1045-1054.`\n\n## 3) Install bcdi\n* Latest version tested : v0.2.8\n* `cd /Packages`\n* `git clone https://github.com/carnisj/bcdi.git`\n* `cd bcdi`\n* `source_p9`\n* `pip install .`\n* cite `DOI: 10.5281/zenodo.3257616`\n\n## 4) Install facet-analyser (Debian 11 only)\n* Send a thank you email to Fred Picca =D\n* `cd /Packages`\n* `git clone https://salsa.debian.org/science-team/facet-analyser.git`\n* `cd facet-analyser`\n* `git checkout`\n* `sudo mk-build-deps -i`\n* Make sure that you have qt installed, for me I had to install `libqt5opengl5-dev` (debian-testing)\n* `debuild -b`\n* if the package creation fail, try to ignore the test in /debian/rules (line 19)\n* `sudo debi`\n* The package is now installed. You can check the locations of its files with the command `dpkg -L facet-analyser`\n* You should see a file named `/usr/lib/x86_64-linux-gnu/paraview-5.9/plugins/FacetAnalyser/FacetAnalyser.so`\n* Now launch `/usr/bin/paraview` (if not installed yet, good luck, refer to `https://www.paraview.org/Wiki/ParaView:Build_And_Install#Installing`)\n* In paraview, go to Tools > Manage Plugins > Load New\n* Here type the path to the plugin that was printed with the `dpkg -L facet-analyser` command.\n* Feel free to add it to `/usr/bin/plugin` so that it is loaded automatically.\n* cite `Grothausmann, R. (2015). Facet Analyser : ParaView plugin for automated facet detection and measurement of interplanar angles of tomographic objects. March.`\n\n\n# To go further ...\n\n## Using `Gwaihir` only as a plotting tool in Jupyter Notebook\n\n\n\n## Quick navigation between `vtk` files in the GUI (outdated but it can give you some ideas)\n\nIt is possible to automate the navigation in the GUI !\n\nHere I have a pandas DataFrame that contains data about my scans, I use to automate the navigation:\n\n```python\nimport time\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport glob\n\ndf = pd.read_csv(\"reconstructions/scans_data.csv\")\n\nGUI.tab_facet.children[3].value = False\nGUI.window.selected_index = 0\ntime.sleep(1)\n\nGUI._list_widgets_init_dir.children[7].value = False\ntime.sleep(1)\n\nscan = 3600\nrow = df[df.scan == scan]\n\nparticle = row.particle.values[0]\ntemp = row.temp_given.values[0]\ncondition = row.condition.values[0]\n\nGUI._list_widgets_init_dir.children[2].value = scan\nGUI._list_widgets_init_dir.children[\n 3].value = f\"/data/id01/inhouse/david/SIXS_June_2021/reconstructions/{temp}/{condition}/{particle}/S{scan}/data/\"\nGUI._list_widgets_init_dir.children[\n 4].value = f\"/data/id01/inhouse/david/SIXS_June_2021/reconstructions/{temp}/{condition}/{particle}/\"\ntime.sleep(1)\nGUI._list_widgets_init_dir.children[7].value = True\n\ntime.sleep(1)\n\nGUI.window.selected_index = 9\n\nGUI.tab_facet.children[3].value = \"load_csv\"\n```\n\n",
"bugtrack_url": null,
"license": "",
"summary": "Python package for BCDI data analysis",
"version": "0.0.6",
"split_keywords": [
"bcdi",
"ipywidgets",
"pynx"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "c58fb6c42fd4975d655b0c2c384abddfe69040449c24495594658b43c80758a4",
"md5": "655e8665a7fe4238b728562b0f82850b",
"sha256": "0fbba5c5172fbac7374ba5ea83dc2330e0f1c454ef17b9e04cfdf5d068f190ed"
},
"downloads": -1,
"filename": "gwaihir-0.0.6.tar.gz",
"has_sig": false,
"md5_digest": "655e8665a7fe4238b728562b0f82850b",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 103538,
"upload_time": "2023-02-05T20:26:02",
"upload_time_iso_8601": "2023-02-05T20:26:02.643294Z",
"url": "https://files.pythonhosted.org/packages/c5/8f/b6c42fd4975d655b0c2c384abddfe69040449c24495594658b43c80758a4/gwaihir-0.0.6.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-02-05 20:26:02",
"github": false,
"gitlab": false,
"bitbucket": false,
"lcname": "gwaihir"
}