# H5CROSS
This package provides basic utilities allowing the comparison of hdf5 type files.
This includes the computation of standard statistics on data arrays such as the mean,
min, max, median and standard deviation.
An interactive mode is enabled through the use of nobvisual (https://pypi.org/project/nobvisual/).
Scatter plot comparative capability is enabled through seaborn and matplotlib.
## Installation
Installation using Pypi:
```bash
> pip install h5cross
```
## Usage
Use the CLI for a basic terminal usage.
```bash
--------------- H5CROSS --------------------
You are now using the Command line interface of h5cross a Python3 helper
to explore and compare hdf5 files, created at CERFACS
(https://cerfacs.fr).
This is a python package currently installed in your python environement.
Options:
--help Show this message and exit.
Commands:
convert Conversion to hdf5 of certain file formats.
diff Compare the content of two hdf5 files and view interactively...
dump Write the content of an hdf5 file into a YAML file.
scatter Scatter plot comparison of two hdf5 files.
stats Compute statistics of arrays from hdf5 file.
tree Print the content of an hdf5 file in terminal.
view Show the content of an hdf5 file interactively with nobvisual.
```
## Possibilities
### Console file structure visualization
The command `>h5cross tree ` prints the structure of an HDF5 file in the console.
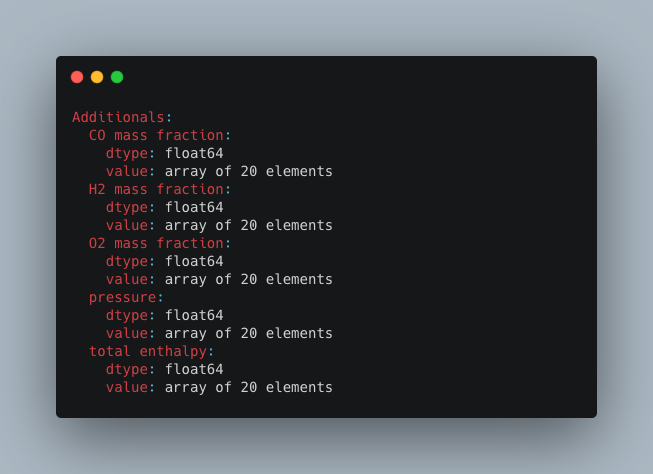
### Statistical information of file
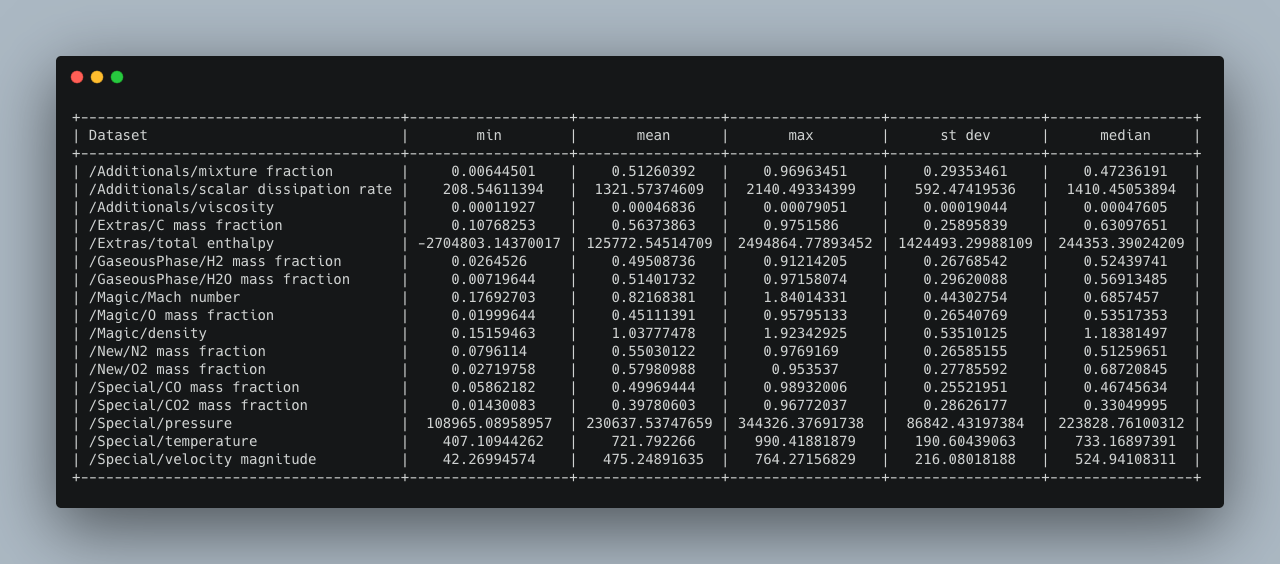
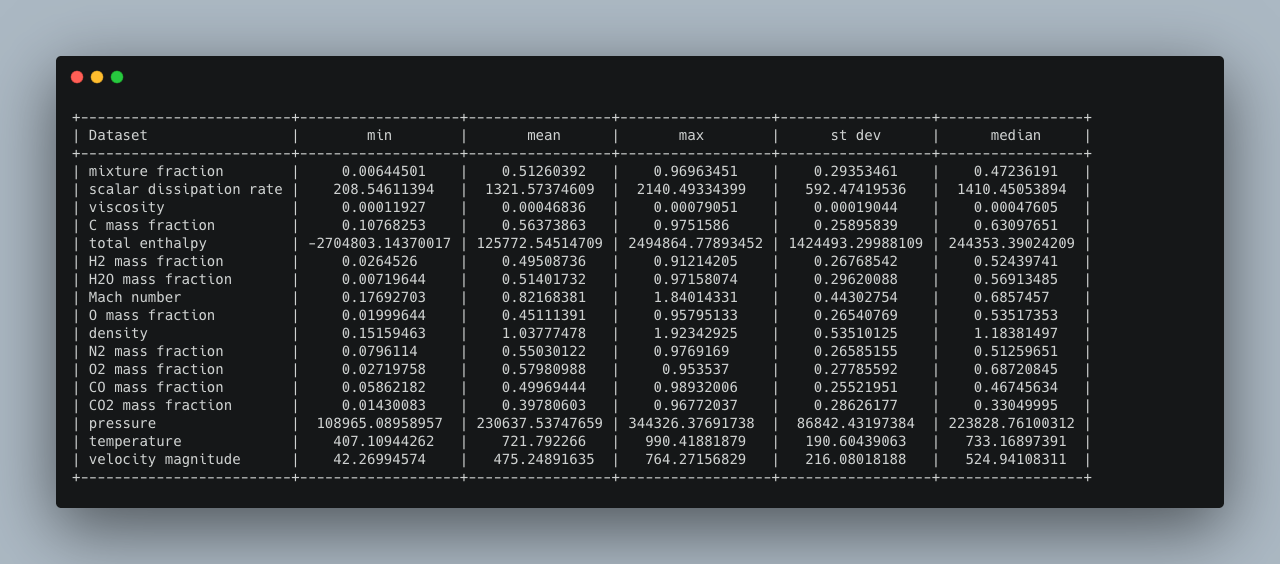
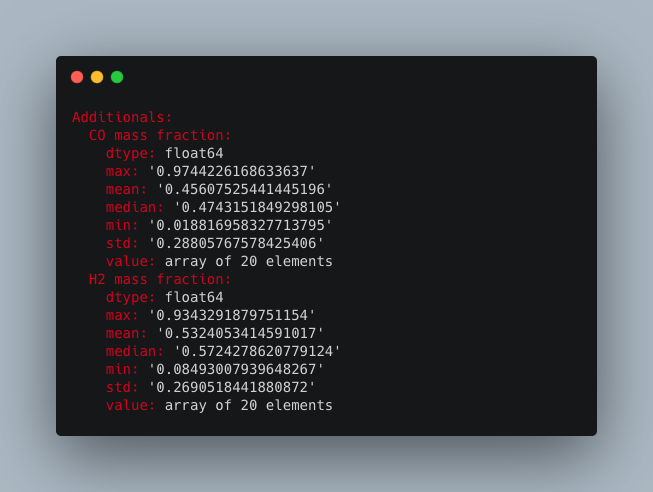
The command `>h5cross stats` computes the mean, min, max, median and standard deviation of every data set for a given HDF5 file. It is then output into the console (can be optionally deactivated). The result can be optionally saved in a YAML format (.yml). A pretty table output is set to default and can be optionally be deactivated in which case a tree output is given. In a pretty table, the full nested path is shown but can be deactivated.
### Save file structure
The command `>h5cross dump` outputs the structure of an HDF5 file in a YAML format (.yml).
### View file structure
The command `>h5cross view` allows an interactive view of the structure of an HDF5 file. It relies on the [nobvisual]( https://pypi.org/project/nobvisual/ ) package designed for the visualisation of nested objects. A temporary .yml file is generated to allow the interactive use.
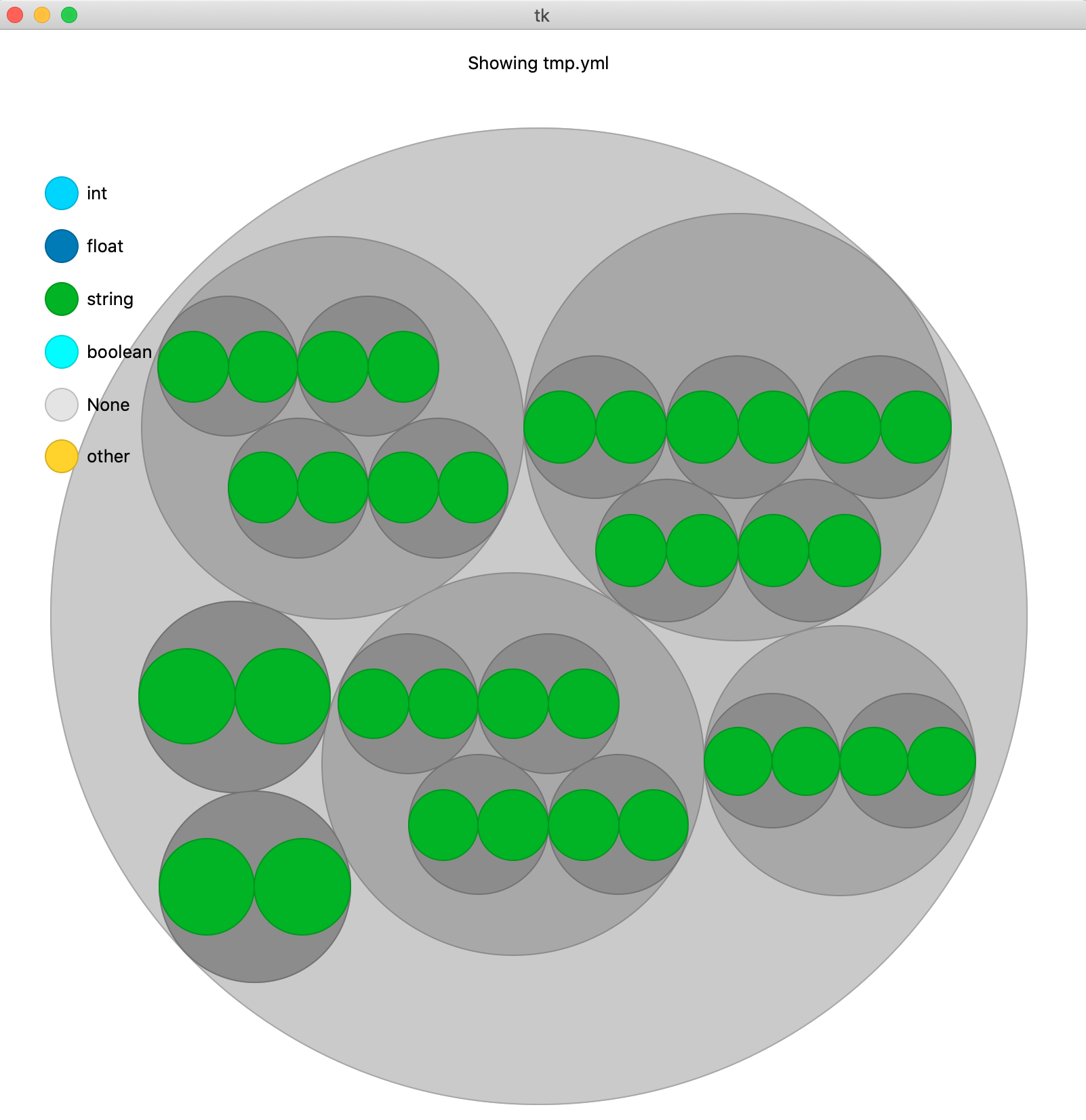
### Compare file structures
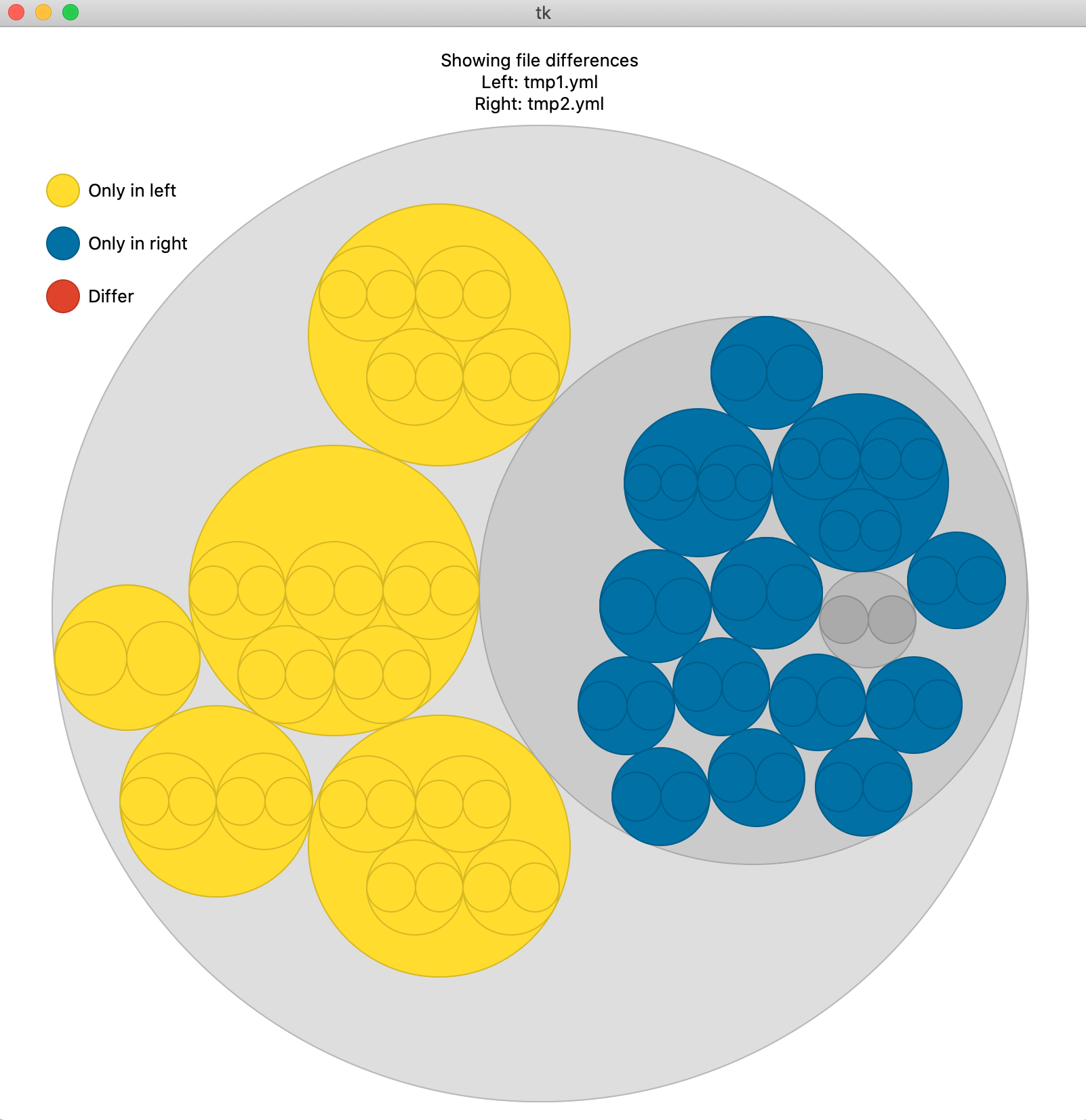
The command `>h5cross diff` compares the structure of two HDF5 files. Similarily to `>h5cross view` it relies on the [nobvisual]( https://pypi.org/project/nobvisual/ ) package. It is optionally possible to add the statistical information of both files in this representation.
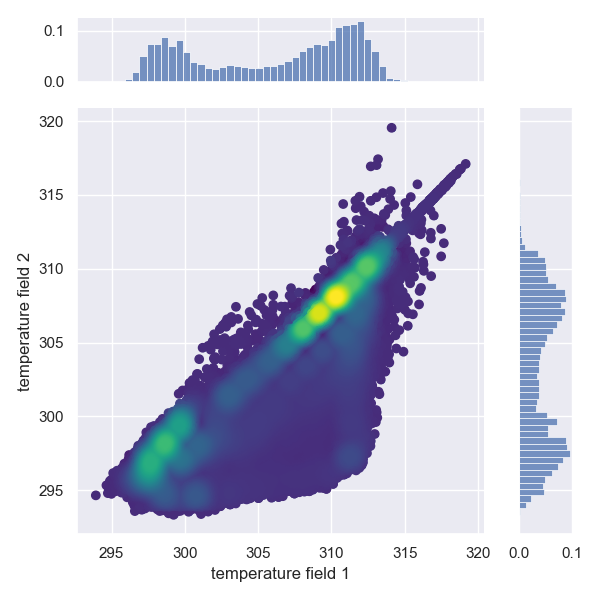
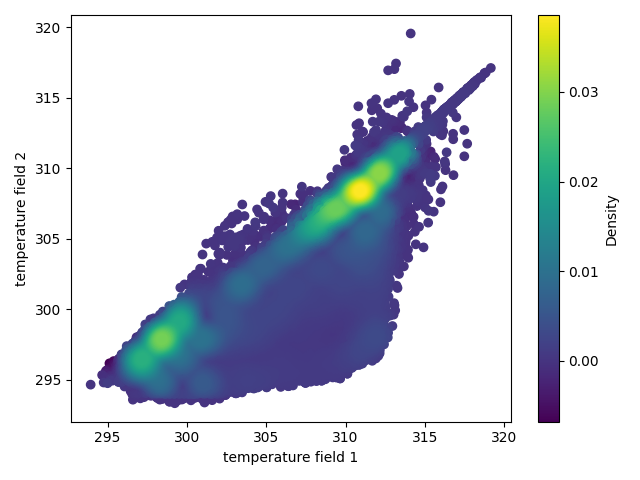
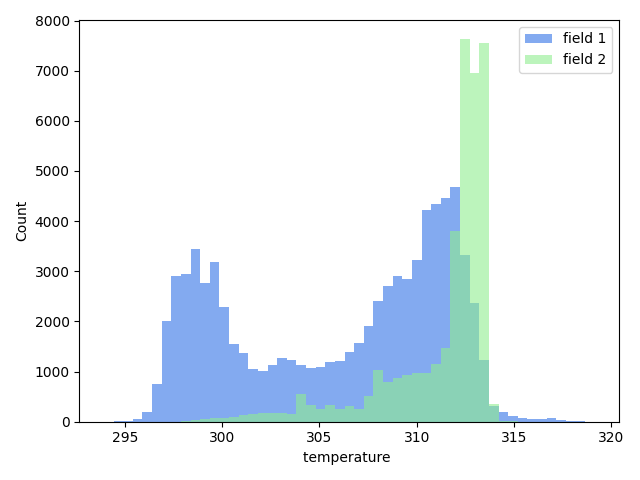
### Compare field data
The command `>h5cross scatter` allows the scatter plot comparison of selected data fields from two HDF5 files. If the data array lengths of each file differ an histogram representation will be given instead.
A minimum requirement is the matplotlib package. By default it uses the seaborn package if available but this setting can be optionally deactivated. The density colormap requires the scipy package, if not available a single color will be used.
### Converting file types to HDF5 file
It is possible to convert certain file formats to HDF5 with `>h5cross convert` .
Currently supported: nek5000, pvtu, vtu, vtk.
## Acknowledgement
h5cross is a service created in the [EXCELLERAT Center Of Excellence](https://www.excellerat.eu/wp/) and is continued as part of the [COEC Center Of Excellence](https://coec-project.eu/). Both projects are funded by the European community.


Raw data
{
"_id": null,
"home_page": "https://gitlab.com/cerfacs/h5cross",
"name": "h5cross",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.7",
"maintainer_email": "",
"keywords": "hdf5,statistics,file conversion,visualization,scientific computing",
"author": "Jimmy-John Hoste, CoopTeam-CERFACS",
"author_email": "coop@cerfacs.fr",
"download_url": "https://files.pythonhosted.org/packages/b1/03/6b28e4b25c80008498a4656fb70b27ccfedcc1a5589ec7cdf53bf8492d9b/h5cross-0.2.3.tar.gz",
"platform": null,
"description": "# H5CROSS\n\nThis package provides basic utilities allowing the comparison of hdf5 type files.\nThis includes the computation of standard statistics on data arrays such as the mean, \nmin, max, median and standard deviation. \nAn interactive mode is enabled through the use of nobvisual (https://pypi.org/project/nobvisual/). \nScatter plot comparative capability is enabled through seaborn and matplotlib. \n\n## Installation\n\nInstallation using Pypi:\n\n```bash\n> pip install h5cross\n```\n\n## Usage\n\nUse the CLI for a basic terminal usage.\n\n```bash\n --------------- H5CROSS --------------------\n\n You are now using the Command line interface of h5cross a Python3 helper\n to explore and compare hdf5 files, created at CERFACS\n (https://cerfacs.fr).\n\n This is a python package currently installed in your python environement.\n\nOptions:\n --help Show this message and exit.\n\nCommands:\n convert Conversion to hdf5 of certain file formats.\n diff Compare the content of two hdf5 files and view interactively...\n dump Write the content of an hdf5 file into a YAML file.\n scatter Scatter plot comparison of two hdf5 files.\n stats Compute statistics of arrays from hdf5 file.\n tree Print the content of an hdf5 file in terminal.\n view Show the content of an hdf5 file interactively with nobvisual.\n\n```\n\n## Possibilities\n\n### Console file structure visualization\n\nThe command `>h5cross tree ` prints the structure of an HDF5 file in the console. \n\n\n### Statistical information of file\n\nThe command `>h5cross stats` computes the mean, min, max, median and standard deviation of every data set for a given HDF5 file. It is then output into the console (can be optionally deactivated). The result can be optionally saved in a YAML format (.yml). A pretty table output is set to default and can be optionally be deactivated in which case a tree output is given. In a pretty table, the full nested path is shown but can be deactivated. \n\n\n\n\n\n\n\n### Save file structure\n\nThe command `>h5cross dump` outputs the structure of an HDF5 file in a YAML format (.yml). \n\n\n\n### View file structure\n\nThe command `>h5cross view` allows an interactive view of the structure of an HDF5 file. It relies on the [nobvisual]( https://pypi.org/project/nobvisual/ ) package designed for the visualisation of nested objects. A temporary .yml file is generated to allow the interactive use. \n\n\n\n### Compare file structures\n\nThe command `>h5cross diff` compares the structure of two HDF5 files. Similarily to `>h5cross view` it relies on the [nobvisual]( https://pypi.org/project/nobvisual/ ) package. It is optionally possible to add the statistical information of both files in this representation. \n\n\n\n### Compare field data \n\nThe command `>h5cross scatter` allows the scatter plot comparison of selected data fields from two HDF5 files. If the data array lengths of each file differ an histogram representation will be given instead. \nA minimum requirement is the matplotlib package. By default it uses the seaborn package if available but this setting can be optionally deactivated. The density colormap requires the scipy package, if not available a single color will be used.\n\n\n\n\n\n\n\n\n### Converting file types to HDF5 file\n\nIt is possible to convert certain file formats to HDF5 with `>h5cross convert` .\nCurrently supported: nek5000, pvtu, vtu, vtk.\n\n## Acknowledgement\n\nh5cross is a service created in the [EXCELLERAT Center Of Excellence](https://www.excellerat.eu/wp/) and is continued as part of the [COEC Center Of Excellence](https://coec-project.eu/). Both projects are funded by the European community.\n\n\n",
"bugtrack_url": null,
"license": "",
"summary": "Helper tools to handle H5 files",
"version": "0.2.3",
"project_urls": {
"Bug Tracker": "https://gitlab.com/cerfacs/h5cross/-/issues",
"Documentation": "https://h5cross.readthedocs.io/en/latest/",
"Homepage": "https://gitlab.com/cerfacs/h5cross"
},
"split_keywords": [
"hdf5",
"statistics",
"file conversion",
"visualization",
"scientific computing"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "82872e515ee49d852b0c488b0c1fb5a80f7285cc1c6fe27c53c531a4f2a20042",
"md5": "41c800763c00a241bc3dcd3db3b1ace0",
"sha256": "00f3858f555852b2c4dbedceda7fd7c39f5e20fba35c8c101ad057ad57aa7133"
},
"downloads": -1,
"filename": "h5cross-0.2.3-py3-none-any.whl",
"has_sig": false,
"md5_digest": "41c800763c00a241bc3dcd3db3b1ace0",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.7",
"size": 26548,
"upload_time": "2023-09-14T14:59:32",
"upload_time_iso_8601": "2023-09-14T14:59:32.583774Z",
"url": "https://files.pythonhosted.org/packages/82/87/2e515ee49d852b0c488b0c1fb5a80f7285cc1c6fe27c53c531a4f2a20042/h5cross-0.2.3-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "b1036b28e4b25c80008498a4656fb70b27ccfedcc1a5589ec7cdf53bf8492d9b",
"md5": "9afc07c775c5107ba9c41a7cad5be963",
"sha256": "729225706e901c1d1f966ab045a5d616a8f4b54e0dac3adc84c383430a001838"
},
"downloads": -1,
"filename": "h5cross-0.2.3.tar.gz",
"has_sig": false,
"md5_digest": "9afc07c775c5107ba9c41a7cad5be963",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.7",
"size": 25905,
"upload_time": "2023-09-14T14:59:35",
"upload_time_iso_8601": "2023-09-14T14:59:35.282654Z",
"url": "https://files.pythonhosted.org/packages/b1/03/6b28e4b25c80008498a4656fb70b27ccfedcc1a5589ec7cdf53bf8492d9b/h5cross-0.2.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-09-14 14:59:35",
"github": false,
"gitlab": true,
"bitbucket": false,
"codeberg": false,
"gitlab_user": "cerfacs",
"gitlab_project": "h5cross",
"lcname": "h5cross"
}