| Name | hdf5view JSON |
| Version |
0.2.5
 JSON
JSON |
| download |
| home_page | None |
| Summary | hdf5view is a python based HDF5 file viewer built on PyQt5/PySide2/PyQt6/PySide6, QtPy, h5py and pyqtgraph. |
| upload_time | 2025-03-18 11:29:52 |
| maintainer | Thomas G. Woodcock |
| docs_url | None |
| author | Martin Swarbrick, Thomas G. Woodcock, The hdf5view Developers |
| requires_python | >=3.6 |
| license | MIT License
Copyright (c) 2019 The hdf5view Developers
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
|
| keywords |
research
data
management
visualization
hdf5
viewer
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
[](https://pypi.python.org/pypi/hdf5view/)
[](https://pypi.python.org/pypi/hdf5view/)
# **hdf5view** <img src="https://raw.githubusercontent.com/tgwoodcock/hdf5view/main/src/hdf5view/resources/images/hdf5view.ico" width="28" />
**Simple Qt/Python based viewer for HDF5 files.**
Features:
- Image tab(s) showing rgb(a) or grayscale images of a selected node:

<br>
- Plot tab(s) showing columns of data in a node:
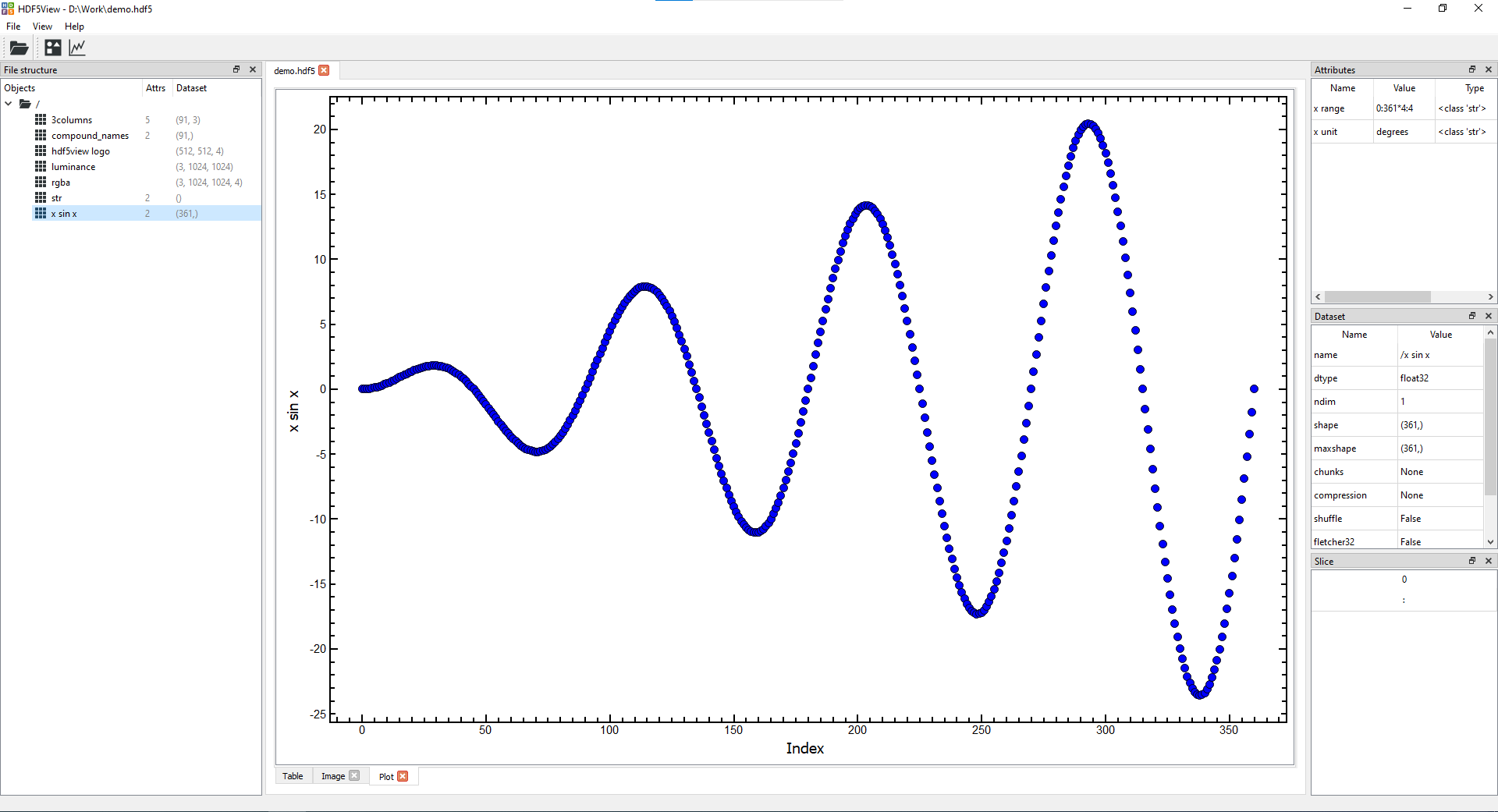
<br>
- Table tab giving a view of the data in the dataset selected:
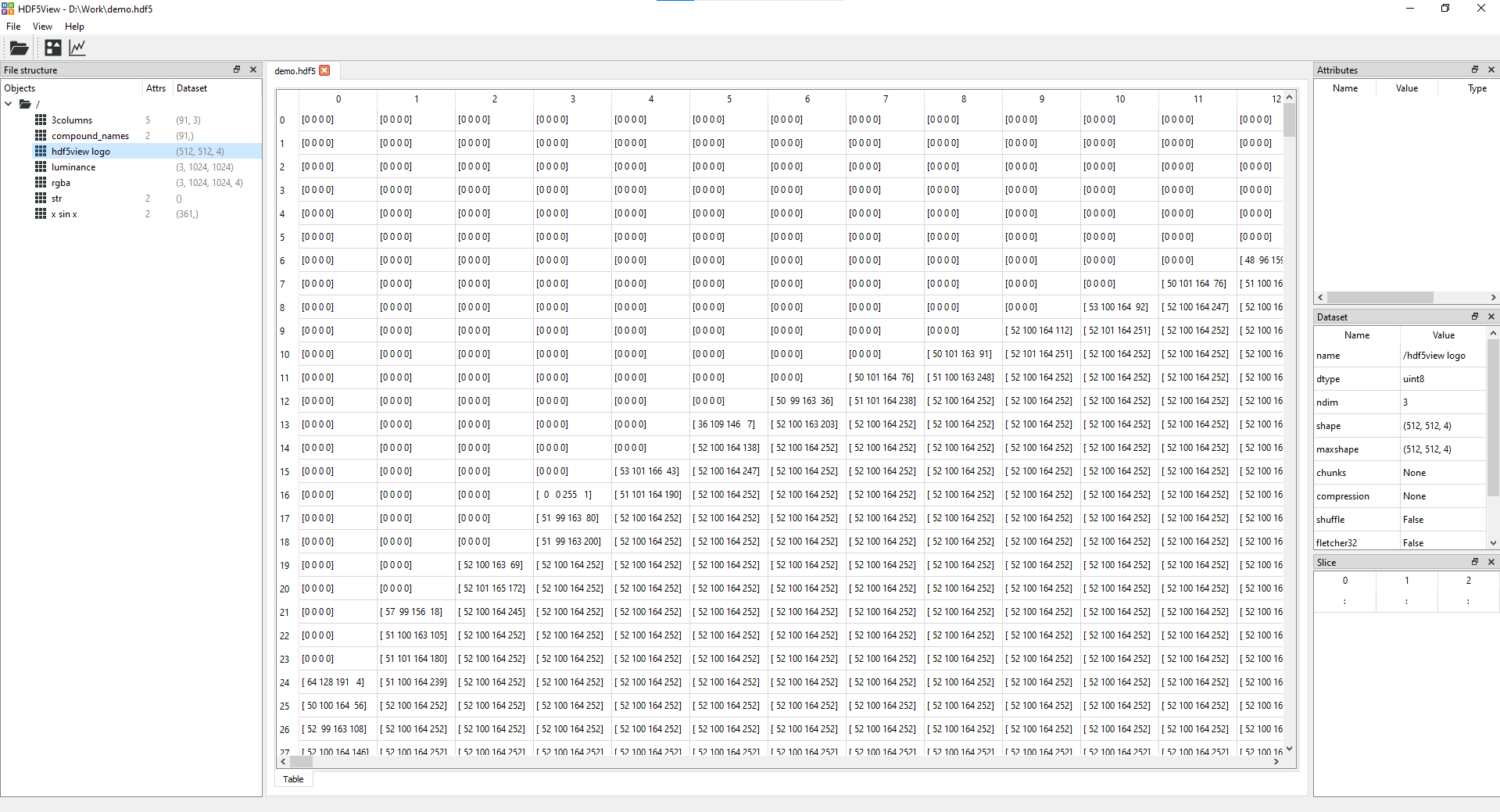
<br>
- File Structure table giving a tree view of the hdf5 file
- Attributes table showing any attributes assigned to a dataset
- Dataset table showing *e.g.* the shape, number of dimensions and data type of a dataset
- Slice table showing which slice of the data is currently displayed (can be set by the user)
- Export images/plots in a variety of formats (image files, data files, hdf5, matplotlib window)
- Datasets are loaded dynamically, so hopefully it should be able to handle HDF5 files of any size and structure.
- Warnings are given when selecting a dataset if loading it would consume more than 30% of the available memory. The user can the opt to abort or continue loading.
<br>
**Why use hdf5view?**
hdf5view is a simple Qt/Python based tool which aims to be easy to use and to allow you to get a view of your data with just a few clicks. If you need to check the structure of an HDF5 file, quickly see what data it contains or choose a dataset for futher analysis in another program, hdf5view may be for you. We don't aim to be able to create or edit HDF5 files and have only minimal possibilties for data analysis other than just viewing it. Other viewers for HDF5 files are available, which may be more suited to your needs, see [related projects](https://tgwoodcock.github.io/hdf5view/user/related_projects.html).
<br>
## Documentation
The documentation for hdf5view is here: [https://tgwoodcock.github.io/hdf5view](https://tgwoodcock.github.io/hdf5view)
<br>
## **1. Installing**
hdf5view is designed to be platform independent and can be installed with `pip`. Please see the [installation guide](https://tgwoodcock.github.io/hdf5view/user/installation.html) for details.
<br>
## **2. Running**
The [usage guide](https://tgwoodcock.github.io/hdf5view/user/basic_usage.html) shows various ways to start hdf5view and open HDF5 files in the application.
#### **Context menu**
A particularly useful way to use hdf5view is to add an entry to the context menu. This way, you can open any HDF5 file with two clicks. Please see the [context menu guide](https://tgwoodcock.github.io/hdf5view/user/basic_usage.html#setting-up-the-context-menu) for instructions on how to set this up.
## **3. Usage**
In the [documentation](https://tgwoodcock.github.io/hdf5view), there are tutorials on how to use
- the [table tab](https://tgwoodcock.github.io/hdf5view/user/table_tab.html)
- the [image tab](https://tgwoodcock.github.io/hdf5view/user/image_tab.html) and
- the [plot tab](https://tgwoodcock.github.io/hdf5view/user/plot_tab.html) to view your data.
<br>
## **4. Testing**
Currently there are no unit tests for this package. The gui has been tested with qtpy=2.2.0, pyqtgraph=0.12.4 and h5py=3.7.0 in combination with pyqt5=5.15.7, pyside2=5.15.2.1, pyqt6=6.3.1 and pyside6=6.3.2, and it works with all of the Qt API bindings.
<br>
## **5. Issues**
If there are any issues, please feel free to use the [issues mechanism on github](https://github.com/tgwoodcock/hdf5view/issues) to get in touch.
<br>
## **6. Contributing**
If you are interested in contributing to the hdf5view project, please see the [contributing guide](https://tgwoodcock.github.io/hdf5view/dev/index.html).
<br>
## **7. Changes**
To see the what has changed in the current version of hdf5view, please have a look at the [changelog](https://tgwoodcock.github.io/hdf5view/changelog.html).
<br>
## **8. Reference**
hdf5view is archived on Zenodo and can be cited using the following doi and associated metadata: [](https://doi.org/10.5281/zenodo.14236673)
<br>
## TODO:
* Implement dynamic loading for files larger than the available memory
* Add tests
* Possibly add 3D rendering, likely based on pyqtgraph
Raw data
{
"_id": null,
"home_page": null,
"name": "hdf5view",
"maintainer": "Thomas G. Woodcock",
"docs_url": null,
"requires_python": ">=3.6",
"maintainer_email": null,
"keywords": "research, data, management, visualization, hdf5, viewer",
"author": "Martin Swarbrick, Thomas G. Woodcock, The hdf5view Developers",
"author_email": null,
"download_url": "https://files.pythonhosted.org/packages/c2/66/0feae8476857e20d9264123de0cf3e36e10dd3cb842547cbf8a8746dd267/hdf5view-0.2.5.tar.gz",
"platform": null,
"description": "[](https://pypi.python.org/pypi/hdf5view/)\r\n[](https://pypi.python.org/pypi/hdf5view/)\r\n\r\n# **hdf5view** <img src=\"https://raw.githubusercontent.com/tgwoodcock/hdf5view/main/src/hdf5view/resources/images/hdf5view.ico\" width=\"28\" /> \r\n\r\n**Simple Qt/Python based viewer for HDF5 files.**\r\n\r\nFeatures:\r\n- Image tab(s) showing rgb(a) or grayscale images of a selected node:\r\n\r\n\r\n\r\n<br>\r\n\r\n- Plot tab(s) showing columns of data in a node:\r\n\r\n\r\n \r\n<br>\r\n\r\n- Table tab giving a view of the data in the dataset selected:\r\n\r\n\r\n\r\n<br>\r\n\r\n- File Structure table giving a tree view of the hdf5 file\r\n- Attributes table showing any attributes assigned to a dataset\r\n- Dataset table showing *e.g.* the shape, number of dimensions and data type of a dataset\r\n- Slice table showing which slice of the data is currently displayed (can be set by the user)\r\n- Export images/plots in a variety of formats (image files, data files, hdf5, matplotlib window)\r\n- Datasets are loaded dynamically, so hopefully it should be able to handle HDF5 files of any size and structure.\r\n- Warnings are given when selecting a dataset if loading it would consume more than 30% of the available memory. The user can the opt to abort or continue loading.\r\n\r\n<br>\r\n\r\n**Why use hdf5view?**\r\n\r\nhdf5view is a simple Qt/Python based tool which aims to be easy to use and to allow you to get a view of your data with just a few clicks. If you need to check the structure of an HDF5 file, quickly see what data it contains or choose a dataset for futher analysis in another program, hdf5view may be for you. We don't aim to be able to create or edit HDF5 files and have only minimal possibilties for data analysis other than just viewing it. Other viewers for HDF5 files are available, which may be more suited to your needs, see [related projects](https://tgwoodcock.github.io/hdf5view/user/related_projects.html).\r\n\r\n<br>\r\n\r\n## Documentation\r\n\r\nThe documentation for hdf5view is here: [https://tgwoodcock.github.io/hdf5view](https://tgwoodcock.github.io/hdf5view)\r\n\r\n<br>\r\n\r\n## **1. Installing**\r\n\r\nhdf5view is designed to be platform independent and can be installed with `pip`. Please see the [installation guide](https://tgwoodcock.github.io/hdf5view/user/installation.html) for details.\r\n\r\n<br>\r\n\r\n## **2. Running**\r\n\r\nThe [usage guide](https://tgwoodcock.github.io/hdf5view/user/basic_usage.html) shows various ways to start hdf5view and open HDF5 files in the application.\r\n\r\n#### **Context menu**\r\n\r\nA particularly useful way to use hdf5view is to add an entry to the context menu. This way, you can open any HDF5 file with two clicks. Please see the [context menu guide](https://tgwoodcock.github.io/hdf5view/user/basic_usage.html#setting-up-the-context-menu) for instructions on how to set this up.\r\n\r\n\r\n## **3. Usage**\r\n\r\nIn the [documentation](https://tgwoodcock.github.io/hdf5view), there are tutorials on how to use\r\n\r\n- the [table tab](https://tgwoodcock.github.io/hdf5view/user/table_tab.html)\r\n- the [image tab](https://tgwoodcock.github.io/hdf5view/user/image_tab.html) and\r\n- the [plot tab](https://tgwoodcock.github.io/hdf5view/user/plot_tab.html) to view your data.\r\n\r\n<br>\r\n\r\n## **4. Testing**\r\n\r\nCurrently there are no unit tests for this package. The gui has been tested with qtpy=2.2.0, pyqtgraph=0.12.4 and h5py=3.7.0 in combination with pyqt5=5.15.7, pyside2=5.15.2.1, pyqt6=6.3.1 and pyside6=6.3.2, and it works with all of the Qt API bindings.\r\n\r\n<br>\r\n\r\n## **5. Issues**\r\n\r\nIf there are any issues, please feel free to use the [issues mechanism on github](https://github.com/tgwoodcock/hdf5view/issues) to get in touch.\r\n\r\n<br>\r\n\r\n## **6. Contributing**\r\n\r\nIf you are interested in contributing to the hdf5view project, please see the [contributing guide](https://tgwoodcock.github.io/hdf5view/dev/index.html).\r\n\r\n<br>\r\n\r\n## **7. Changes**\r\n\r\nTo see the what has changed in the current version of hdf5view, please have a look at the [changelog](https://tgwoodcock.github.io/hdf5view/changelog.html).\r\n\r\n<br>\r\n\r\n## **8. Reference**\r\n\r\nhdf5view is archived on Zenodo and can be cited using the following doi and associated metadata: [](https://doi.org/10.5281/zenodo.14236673)\r\n\r\n<br>\r\n\r\n## TODO:\r\n\r\n* Implement dynamic loading for files larger than the available memory\r\n* Add tests\r\n* Possibly add 3D rendering, likely based on pyqtgraph\r\n",
"bugtrack_url": null,
"license": "MIT License\r\n \r\n Copyright (c) 2019 The hdf5view Developers\r\n \r\n Permission is hereby granted, free of charge, to any person obtaining a copy\r\n of this software and associated documentation files (the \"Software\"), to deal\r\n in the Software without restriction, including without limitation the rights\r\n to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\r\n copies of the Software, and to permit persons to whom the Software is\r\n furnished to do so, subject to the following conditions:\r\n \r\n The above copyright notice and this permission notice shall be included in all\r\n copies or substantial portions of the Software.\r\n \r\n THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\r\n IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\r\n FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\r\n AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\r\n LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\r\n OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\r\n SOFTWARE.\r\n ",
"summary": "hdf5view is a python based HDF5 file viewer built on PyQt5/PySide2/PyQt6/PySide6, QtPy, h5py and pyqtgraph.",
"version": "0.2.5",
"project_urls": {
"Bug Tracker": "https://github.com/tgwoodcock/hdf5view/issues",
"Changelog": "https://github.com/tgwoodcock/hdf5view/blob/main/CHANGELOG.rst",
"Documentation": "https://tgwoodcock.github.io/hdf5view",
"Source": "https://github.com/tgwoodcock/hdf5view/"
},
"split_keywords": [
"research",
" data",
" management",
" visualization",
" hdf5",
" viewer"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "2a9745408a283a6cc800995a260380debe3cda6fc1b98c57b89df2c9e0db9b93",
"md5": "96b0a4e2ad76c3c59524dbf4727917f3",
"sha256": "821a4653a1383a6fbf5627cd30df520ce8d1a0c4beae2e99c544edb94d06003d"
},
"downloads": -1,
"filename": "hdf5view-0.2.5-py3-none-any.whl",
"has_sig": false,
"md5_digest": "96b0a4e2ad76c3c59524dbf4727917f3",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 56880,
"upload_time": "2025-03-18T11:29:50",
"upload_time_iso_8601": "2025-03-18T11:29:50.357833Z",
"url": "https://files.pythonhosted.org/packages/2a/97/45408a283a6cc800995a260380debe3cda6fc1b98c57b89df2c9e0db9b93/hdf5view-0.2.5-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "c2660feae8476857e20d9264123de0cf3e36e10dd3cb842547cbf8a8746dd267",
"md5": "d182a7ba4bd2d4f84648062158acd13a",
"sha256": "4279d50546eff06152c712455b4fccec5996cbfdaf5b0fdbdb8abcf31c4f442a"
},
"downloads": -1,
"filename": "hdf5view-0.2.5.tar.gz",
"has_sig": false,
"md5_digest": "d182a7ba4bd2d4f84648062158acd13a",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 57263,
"upload_time": "2025-03-18T11:29:52",
"upload_time_iso_8601": "2025-03-18T11:29:52.011297Z",
"url": "https://files.pythonhosted.org/packages/c2/66/0feae8476857e20d9264123de0cf3e36e10dd3cb842547cbf8a8746dd267/hdf5view-0.2.5.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-03-18 11:29:52",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "tgwoodcock",
"github_project": "hdf5view",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "hdf5view"
}
