<img alt="IFermi logo" src="https://raw.githubusercontent.com/fermisurfaces/IFermi/main/docs/_static/logo2-01.png" height="150px">
--------
[📖 **Official Documentation** 📖](https://fermisurfaces.github.io/IFermi)
[🙋 **Support Forum** 🙋](https://matsci.org/c/ifermi/)
[📝 **JOSS Paper** 📝](https://doi.org/10.21105/joss.03089)
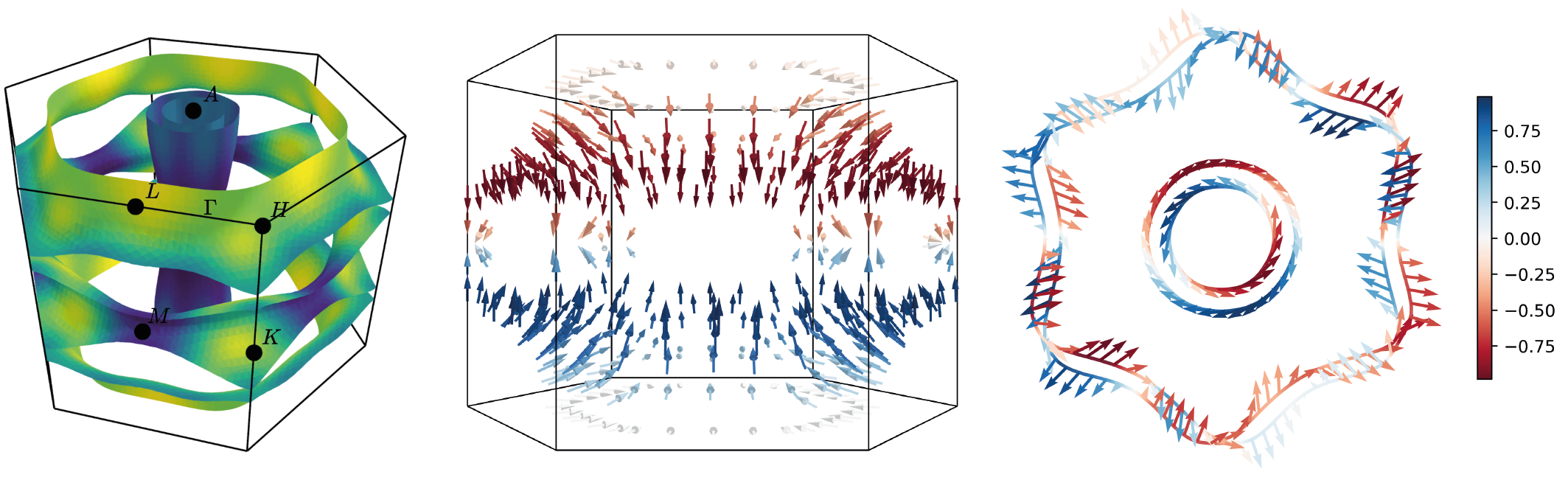
IFermi is a Python (3.9+) library and set of command-line tools for the generation,
analysis, and visualisation of Fermi surfaces and Fermi slices. The goal of the library
is to provide fully featured FermiSurface and FermiSlice objects that allow for easy
manipulation and analysis. The main features include:
- Interpolation of electronic band structures onto dense k-point meshes.
- Extraction of Fermi surfaces and Fermi slices from electronic band structures.
- Projection of arbitrary properties onto Fermi surfaces and Fermi slices.
- Tools to calculate Fermi surface dimensionality, orientation, and averaged projections,
including Fermi velocities.
- Interactive visualisation of Fermi surfaces and slices, with support for
[mayavi](https://docs.enthought.com/mayavi/mayavi/), [plotly](https://plot.ly/) and
[matplotlib](https://matplotlib.org).
- Generation and visualisation of spin-texture.
IFermi's command-line tools only work with VASP calculations but support for additional
DFT packages will be added in the future.
## Quick start
The [online documentation](https://fermisurfaces.github.io/IFermi/cli.html) provides a full
description of the available command-line options.
### Analysis
Fermi surface properties, including dimensionality and orientation can be extracted
from a vasprun.xml file using:
```bash
ifermi info --property velocity
```
```
Fermi Surface Summary
=====================
# surfaces: 5
Area: 32.75 Å⁻²
Avg velocity: 9.131e+05 m/s
Isosurfaces
~~~~~~~~~~~
Band Area [Å⁻²] Velocity avg [m/s] Dimensionality Orientation
------ ------------ -------------------- ---------------- -------------
6 1.944 7.178e+05 2D (0, 0, 1)
7 4.370 9.092e+05 quasi-2D (0, 0, 1)
7 2.961 5.880e+05 2D (0, 0, 1)
8 3.549 1.105e+06 quasi-2D (0, 0, 1)
8 3.549 1.105e+06 quasi-2D (0, 0, 1)
```
### Visualisation
Three-dimensional Fermi surfaces can be visualized from a `vasprun.xml` file using:
```bash
ifermi plot
```
The two-dimensional slice of a Fermi surface along the plane specified by the miller
indices (j k l) and distance d can be plotted from a `vasprun.xml` file using:
```bash
ifermi plot --slice j k l d
```
### Python library
The `ifermi` command line tools are build on the IFermi Python library. Here is an
example of how to load DFT calculation outputs, interpolate the energies onto a dense mesh,
generate a Fermi surface, calculate Fermi surface properties, and visualise the surface.
A more complete summary of the API is given in the [API introduction page](https://fermisurfaces.github.io/IFermi/introduction_to_ifermi.html)
and in the [API Reference page](https://fermisurfaces.github.io/IFermi/reference.html) in the documentation.
```python
from pymatgen.io.vasp.outputs import Vasprun
from ifermi.surface import FermiSurface
from ifermi.interpolate import FourierInterpolator
from ifermi.plot import FermiSlicePlotter, FermiSurfacePlotter, save_plot, show_plot
from ifermi.kpoints import kpoints_from_bandstructure
# load VASP calculation outputs
vr = Vasprun("vasprun.xml")
bs = vr.get_band_structure()
# interpolate the energies onto a dense k-point mesh
interpolator = FourierInterpolator(bs)
dense_bs, velocities = interpolator.interpolate_bands(return_velocities=True)
# generate the Fermi surface and calculate the dimensionality
fs = FermiSurface.from_band_structure(
dense_bs, mu=0.0, wigner_seitz=True, calculate_dimensionality=True
)
# generate the Fermi surface and calculate the group velocity at the
# center of each triangular face
dense_kpoints = kpoints_from_bandstructure(dense_bs)
fs = FermiSurface.from_band_structure(
dense_bs, mu=0.0, wigner_seitz=True, calculate_dimensionality=True,
property_data=velocities, property_kpoints=dense_kpoints
)
# number of isosurfaces in the Fermi surface
fs.n_surfaces
# number of isosurfaces for each Spin channel
fs.n_surfaces_per_spin
# the total area of the Fermi surface
fs.area
# the area of each isosurface
fs.area_surfaces
# loop over all isosurfaces and check their properties
# the isosurfaces are given as a list for each spin channel
for spin, isosurfaces in fs.isosurfaces.items():
for isosurface in isosurfaces:
# the dimensionality (does the surface cross periodic boundaries)
isosurface.dimensionality
# what is the orientation
isosurface.orientation
# does the surface have face properties
isosurface.has_properties
# calculate the norms of the properties
isosurface.properties_norms
# calculate scalar projection of properties on to [0 0 1] vector
isosurface.scalar_projection((0, 0, 1))
# uniformly sample the surface faces to a consistent density
isosurface.sample_uniform(0.1)
# plot the Fermi surface
fs_plotter = FermiSurfacePlotter(fs)
plot = fs_plotter.get_plot()
# generate Fermi slice along the (0 0 1) plane going through the Γ-point.
fermi_slice = fs.get_fermi_slice((0, 0, 1))
# number of isolines in the slice
fermi_slice.n_lines
# do the lines have segment properties
fermi_slice.has_properties
# plot slice
slice_plotter = FermiSlicePlotter(fermi_slice)
plot = slice_plotter.get_plot()
save_plot(plot, "fermi-slice.png") # saves the plot to a file
show_plot(plot) # displays an interactive plot
```
## Citing IFermi
If you find IFermi useful, please encourage its development by citing the following
[paper](https://doi.org/10.21105/joss.03089) in your research output:
```
Ganose, A. M., Searle, A., Jain, A., Griffin, S. M., IFermi: A python library for Fermi
surface generation and analysis. Journal of Open Source Software, 2021, 6 (59), 3089
```
## Installation
The recommended way to install IFermi is in a conda environment.
```bash
conda create --name ifermi pip cmake numpy
conda activate ifermi
conda install -c conda-forge pymatgen boltztrap2 pyfftw
pip install ifermi
````
IFermi is currently compatible with Python 3.9+ and relies on a number of
open-source python packages, specifically:
- [pymatgen](http://pymatgen.org) for parsing DFT calculation outputs.
- [BoltzTrap2](https://gitlab.com/sousaw/BoltzTraP2) for band structure interpolation.
- [trimesh](https://trimsh.org/) for manipulating isosurfaces.
- [matplotlib](https://matplotlib.org), [mayavi](https://docs.enthought.com/mayavi/mayavi/), and [plotly](https://plot.ly/) for three-dimensional plotting.
### Running tests
The integration tests can be run to ensure IFermi has been installed correctly. First
download the IFermi source and install the test requirements.
```
git clone https://github.com/fermisurfaces/IFermi.git
cd IFermi
pip install .[tests]
```
The tests can be run in the IFermi folder using:
```bash
pytest
```
## Need Help?
Ask questions about the IFermi Python API and command-line tools on the [IFermi
support forum](https://matsci.org/c/ifermi).
If you've found an issue with IFermi, please submit a bug report
[here](https://github.com/fermisurfaces/IFermi/issues).
## What’s new?
Track changes to IFermi through the
[changelog](https://fermisurfaces.github.io/IFermi/changelog.html).
## Contributing
We greatly appreciate any contributions in the form of a pull request.
Additional information on contributing to IFermi can be found [here](https://fermisurfaces.github.io/IFermi/contributing.html).
We maintain a list of all contributors [here](https://fermisurfaces.github.io/IFermi/contributors.html).
## License
IFermi is made available under the MIT License (see LICENSE file).
## Acknowledgements
Developed by Amy Searle and Alex Ganose.
Sinéad Griffin designed and led the project.
Raw data
{
"_id": null,
"home_page": null,
"name": "ifermi",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.9",
"maintainer_email": null,
"keywords": "fermi-surface, pymatgen, dft, vasp, band, materials-science",
"author": null,
"author_email": "Amy Searle <amyjadesearle@gmail.com>",
"download_url": "https://files.pythonhosted.org/packages/84/03/563a91a624cbf37beb0aaf64f07eee8accdb5c26e7aa8994c6de24aa3468/ifermi-0.3.5.tar.gz",
"platform": null,
"description": "<img alt=\"IFermi logo\" src=\"https://raw.githubusercontent.com/fermisurfaces/IFermi/main/docs/_static/logo2-01.png\" height=\"150px\">\n\n--------\n\n[\ud83d\udcd6 **Official Documentation** \ud83d\udcd6](https://fermisurfaces.github.io/IFermi)\n\n[\ud83d\ude4b **Support Forum** \ud83d\ude4b](https://matsci.org/c/ifermi/)\n\n[\ud83d\udcdd **JOSS Paper** \ud83d\udcdd](https://doi.org/10.21105/joss.03089)\n\nIFermi is a Python (3.9+) library and set of command-line tools for the generation,\nanalysis, and visualisation of Fermi surfaces and Fermi slices. The goal of the library\nis to provide fully featured FermiSurface and FermiSlice objects that allow for easy\nmanipulation and analysis. The main features include:\n\n- Interpolation of electronic band structures onto dense k-point meshes.\n- Extraction of Fermi surfaces and Fermi slices from electronic band structures.\n- Projection of arbitrary properties onto Fermi surfaces and Fermi slices.\n- Tools to calculate Fermi surface dimensionality, orientation, and averaged projections,\n including Fermi velocities.\n- Interactive visualisation of Fermi surfaces and slices, with support for\n [mayavi](https://docs.enthought.com/mayavi/mayavi/), [plotly](https://plot.ly/) and\n [matplotlib](https://matplotlib.org).\n- Generation and visualisation of spin-texture.\n\nIFermi's command-line tools only work with VASP calculations but support for additional\nDFT packages will be added in the future.\n\n\n\n## Quick start\n\nThe [online documentation](https://fermisurfaces.github.io/IFermi/cli.html) provides a full\ndescription of the available command-line options.\n\n### Analysis\n\nFermi surface properties, including dimensionality and orientation can be extracted\nfrom a vasprun.xml file using:\n\n```bash\nifermi info --property velocity\n```\n\n```\nFermi Surface Summary\n=====================\n\n # surfaces: 5\n Area: 32.75 \u00c5\u207b\u00b2\n Avg velocity: 9.131e+05 m/s\n\nIsosurfaces\n~~~~~~~~~~~\n\n Band Area [\u00c5\u207b\u00b2] Velocity avg [m/s] Dimensionality Orientation\n ------ ------------ -------------------- ---------------- -------------\n 6 1.944 7.178e+05 2D (0, 0, 1)\n 7 4.370 9.092e+05 quasi-2D (0, 0, 1)\n 7 2.961 5.880e+05 2D (0, 0, 1)\n 8 3.549 1.105e+06 quasi-2D (0, 0, 1)\n 8 3.549 1.105e+06 quasi-2D (0, 0, 1)\n```\n\n### Visualisation\n\nThree-dimensional Fermi surfaces can be visualized from a `vasprun.xml` file using:\n\n```bash\nifermi plot\n```\n\nThe two-dimensional slice of a Fermi surface along the plane specified by the miller\nindices (j k l) and distance d can be plotted from a `vasprun.xml` file using:\n\n```bash\nifermi plot --slice j k l d\n```\n\n### Python library\n\nThe `ifermi` command line tools are build on the IFermi Python library. Here is an\nexample of how to load DFT calculation outputs, interpolate the energies onto a dense mesh,\ngenerate a Fermi surface, calculate Fermi surface properties, and visualise the surface.\nA more complete summary of the API is given in the [API introduction page](https://fermisurfaces.github.io/IFermi/introduction_to_ifermi.html)\nand in the [API Reference page](https://fermisurfaces.github.io/IFermi/reference.html) in the documentation.\n\n```python\nfrom pymatgen.io.vasp.outputs import Vasprun\nfrom ifermi.surface import FermiSurface\nfrom ifermi.interpolate import FourierInterpolator\nfrom ifermi.plot import FermiSlicePlotter, FermiSurfacePlotter, save_plot, show_plot\nfrom ifermi.kpoints import kpoints_from_bandstructure\n\n# load VASP calculation outputs\nvr = Vasprun(\"vasprun.xml\")\nbs = vr.get_band_structure()\n\n# interpolate the energies onto a dense k-point mesh\ninterpolator = FourierInterpolator(bs)\ndense_bs, velocities = interpolator.interpolate_bands(return_velocities=True)\n\n# generate the Fermi surface and calculate the dimensionality\nfs = FermiSurface.from_band_structure(\n dense_bs, mu=0.0, wigner_seitz=True, calculate_dimensionality=True\n)\n\n# generate the Fermi surface and calculate the group velocity at the\n# center of each triangular face\ndense_kpoints = kpoints_from_bandstructure(dense_bs)\nfs = FermiSurface.from_band_structure(\n dense_bs, mu=0.0, wigner_seitz=True, calculate_dimensionality=True,\n property_data=velocities, property_kpoints=dense_kpoints\n)\n\n# number of isosurfaces in the Fermi surface\nfs.n_surfaces\n\n# number of isosurfaces for each Spin channel\nfs.n_surfaces_per_spin\n\n# the total area of the Fermi surface\nfs.area\n\n# the area of each isosurface\nfs.area_surfaces\n\n# loop over all isosurfaces and check their properties\n# the isosurfaces are given as a list for each spin channel\nfor spin, isosurfaces in fs.isosurfaces.items():\n for isosurface in isosurfaces:\n # the dimensionality (does the surface cross periodic boundaries)\n isosurface.dimensionality\n\n # what is the orientation\n isosurface.orientation\n\n # does the surface have face properties\n isosurface.has_properties\n\n # calculate the norms of the properties\n isosurface.properties_norms\n\n # calculate scalar projection of properties on to [0 0 1] vector\n isosurface.scalar_projection((0, 0, 1))\n\n # uniformly sample the surface faces to a consistent density\n isosurface.sample_uniform(0.1)\n\n# plot the Fermi surface\nfs_plotter = FermiSurfacePlotter(fs)\nplot = fs_plotter.get_plot()\n\n# generate Fermi slice along the (0 0 1) plane going through the \u0393-point.\nfermi_slice = fs.get_fermi_slice((0, 0, 1))\n\n# number of isolines in the slice\nfermi_slice.n_lines\n\n# do the lines have segment properties\nfermi_slice.has_properties\n\n# plot slice\nslice_plotter = FermiSlicePlotter(fermi_slice)\nplot = slice_plotter.get_plot()\n\nsave_plot(plot, \"fermi-slice.png\") # saves the plot to a file\nshow_plot(plot) # displays an interactive plot\n```\n\n## Citing IFermi\n\nIf you find IFermi useful, please encourage its development by citing the following\n[paper](https://doi.org/10.21105/joss.03089) in your research output:\n\n```\nGanose, A. M., Searle, A., Jain, A., Griffin, S. M., IFermi: A python library for Fermi\nsurface generation and analysis. Journal of Open Source Software, 2021, 6 (59), 3089\n```\n\n\n## Installation\n\nThe recommended way to install IFermi is in a conda environment.\n\n```bash\nconda create --name ifermi pip cmake numpy\nconda activate ifermi\nconda install -c conda-forge pymatgen boltztrap2 pyfftw\npip install ifermi\n````\n\nIFermi is currently compatible with Python 3.9+ and relies on a number of\nopen-source python packages, specifically:\n\n- [pymatgen](http://pymatgen.org) for parsing DFT calculation outputs.\n- [BoltzTrap2](https://gitlab.com/sousaw/BoltzTraP2) for band structure interpolation.\n- [trimesh](https://trimsh.org/) for manipulating isosurfaces.\n- [matplotlib](https://matplotlib.org), [mayavi](https://docs.enthought.com/mayavi/mayavi/), and [plotly](https://plot.ly/) for three-dimensional plotting.\n\n### Running tests\n\nThe integration tests can be run to ensure IFermi has been installed correctly. First\ndownload the IFermi source and install the test requirements.\n\n```\ngit clone https://github.com/fermisurfaces/IFermi.git\ncd IFermi\npip install .[tests]\n```\n\nThe tests can be run in the IFermi folder using:\n\n```bash\npytest\n```\n\n## Need Help?\n\nAsk questions about the IFermi Python API and command-line tools on the [IFermi\nsupport forum](https://matsci.org/c/ifermi).\nIf you've found an issue with IFermi, please submit a bug report\n[here](https://github.com/fermisurfaces/IFermi/issues).\n\n## What\u2019s new?\n\nTrack changes to IFermi through the\n[changelog](https://fermisurfaces.github.io/IFermi/changelog.html).\n\n## Contributing\n\nWe greatly appreciate any contributions in the form of a pull request.\nAdditional information on contributing to IFermi can be found [here](https://fermisurfaces.github.io/IFermi/contributing.html).\nWe maintain a list of all contributors [here](https://fermisurfaces.github.io/IFermi/contributors.html).\n\n## License\n\nIFermi is made available under the MIT License (see LICENSE file).\n\n## Acknowledgements\n\nDeveloped by Amy Searle and Alex Ganose.\nSin\u00e9ad Griffin designed and led the project.\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Fermi surface plotting tool from DFT output",
"version": "0.3.5",
"project_urls": {
"changelog": "https://github.com/fermisurfaces/IFermi/blob/main/CHANGELOG.md",
"documentation": "https://fermisurfaces.github.io/IFermi/",
"homepage": "https://fermisurfaces.github.io/IFermi/",
"repository": "https://github.com/fermisurfaces/IFermi"
},
"split_keywords": [
"fermi-surface",
" pymatgen",
" dft",
" vasp",
" band",
" materials-science"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "37f38cb7eda2e1d9dee72ec4621e386d0113eec0828dcd4c0af8decf61912933",
"md5": "9f8950bb0f82f70eead5152173414b1d",
"sha256": "f56d1429cfd0792398c36ca184ad4e967575bb10a74f8b70d79f62f7a1476cc7"
},
"downloads": -1,
"filename": "ifermi-0.3.5-py3-none-any.whl",
"has_sig": false,
"md5_digest": "9f8950bb0f82f70eead5152173414b1d",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.9",
"size": 50030,
"upload_time": "2024-08-20T11:09:36",
"upload_time_iso_8601": "2024-08-20T11:09:36.042871Z",
"url": "https://files.pythonhosted.org/packages/37/f3/8cb7eda2e1d9dee72ec4621e386d0113eec0828dcd4c0af8decf61912933/ifermi-0.3.5-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "8403563a91a624cbf37beb0aaf64f07eee8accdb5c26e7aa8994c6de24aa3468",
"md5": "34afa2501a9fc9ee1e1fee2b0842876d",
"sha256": "4ce71dd592127a875da11c3d325a94f6409f26459b98c53a1fbbd31ee0d76916"
},
"downloads": -1,
"filename": "ifermi-0.3.5.tar.gz",
"has_sig": false,
"md5_digest": "34afa2501a9fc9ee1e1fee2b0842876d",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.9",
"size": 50741,
"upload_time": "2024-08-20T11:09:38",
"upload_time_iso_8601": "2024-08-20T11:09:38.162535Z",
"url": "https://files.pythonhosted.org/packages/84/03/563a91a624cbf37beb0aaf64f07eee8accdb5c26e7aa8994c6de24aa3468/ifermi-0.3.5.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-08-20 11:09:38",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "fermisurfaces",
"github_project": "IFermi",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "ifermi"
}
