# ipycytoscape
[](https://github.com/cytoscape/ipycytoscape/actions/workflows/test.yml) [](https://ipycytoscape.readthedocs.io/en/latest/?badge=master) [](https://stackoverflow.com/questions/tagged/ipycytoscape) [](https://gitter.im/QuantStack/Lobby?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge)
A widget enabling interactive graph visualization with [cytoscape.js](https://js.cytoscape.org/) in JupyterLab and the Jupyter notebook.
Try it out using binder: [](https://mybinder.org/v2/gh/cytoscape/ipycytoscape/HEAD?filepath=examples) or install and try out the [examples](examples).
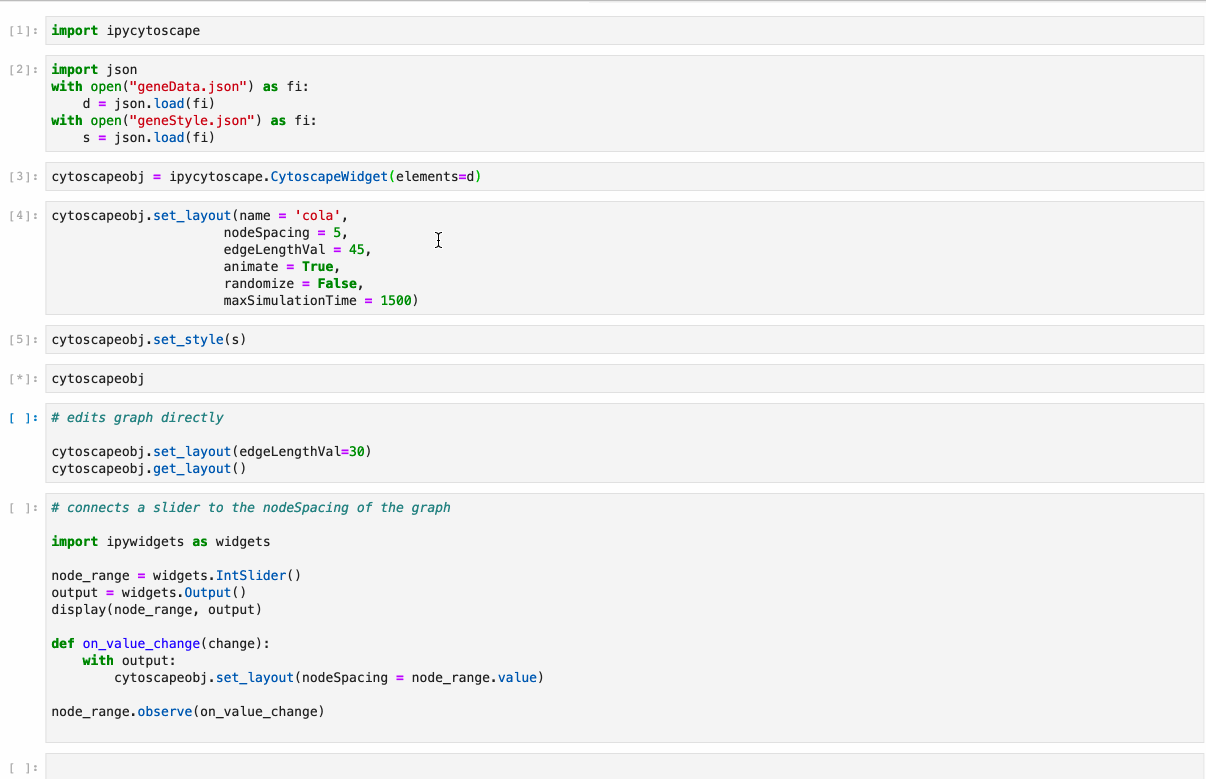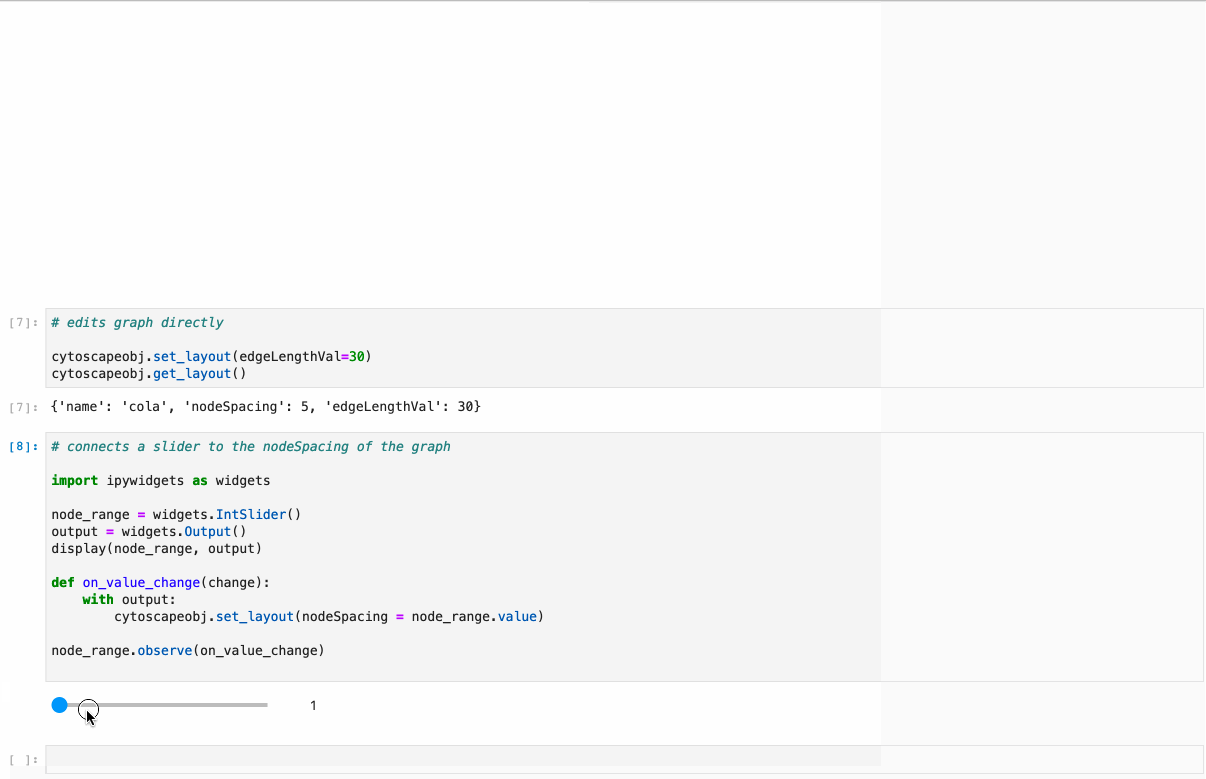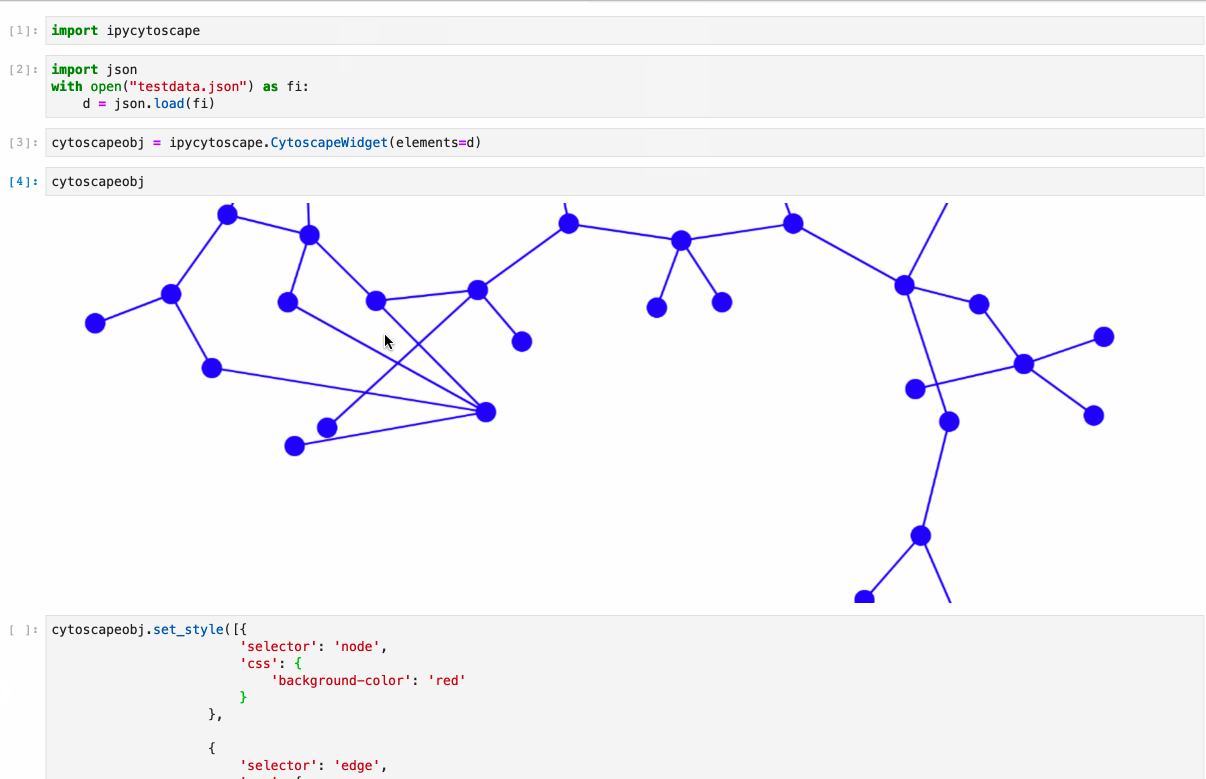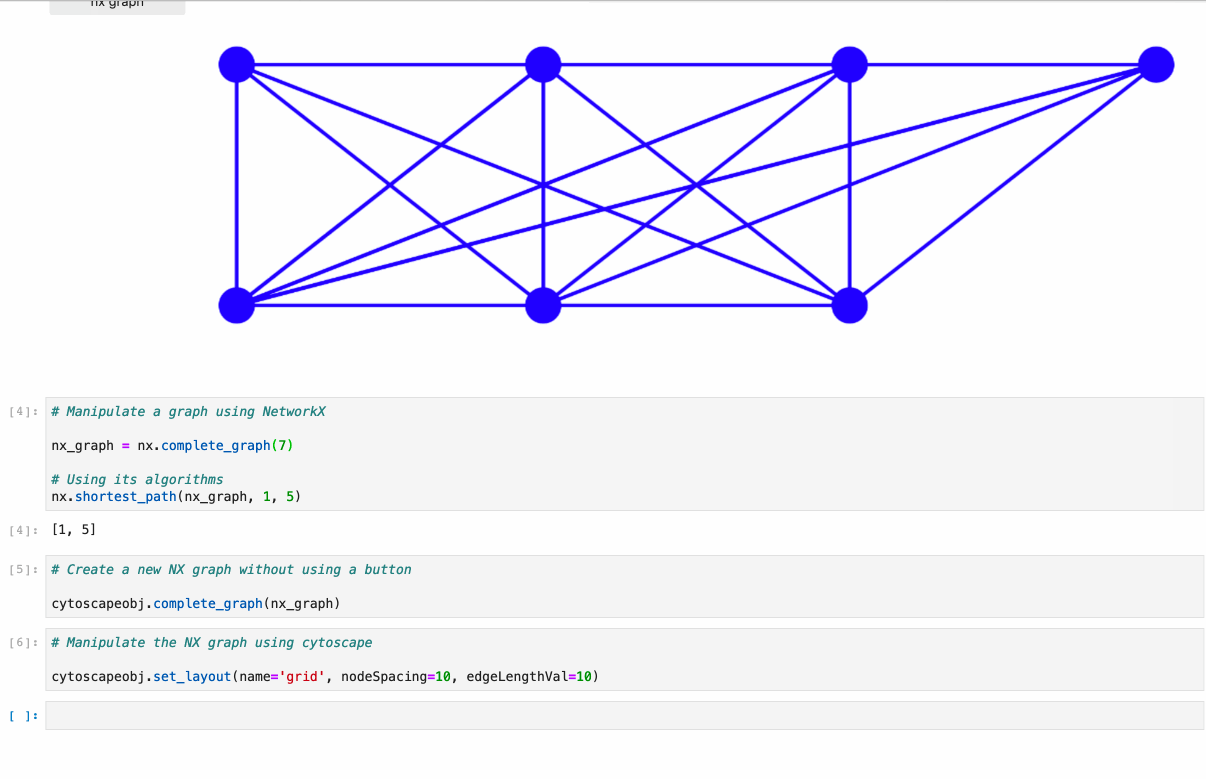
#### Supports:
* Conversion from NetworkX see [example1](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Test%20NetworkX%20methods.ipynb), [example2](https://github.com/cytoscape/ipycytoscape/blob/master/examples/NetworkX%20Example.ipynb)
* Conversion from Pandas DataFrame see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/pandas.ipynb)
* Conversion from neo4j see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Neo4j_Example.ipynb)
## Installation
With `mamba`:
```
mamba install -c conda-forge ipycytoscape
```
With `conda`:
```
conda install -c conda-forge ipycytoscape
```
With `pip`:
```bash
pip install ipycytoscape
```
### Pandas installation
You can install the Pandas dependencies for `ipycytoscape` with pip:
```
pip install pandas
```
Or conda-forge:
```
mamba install pandas
```
### Neo4j installation
You can install the neo4j dependencies for `ipycytoscape` with pip:
```
pip install -e ".[neo4j]"
```
Or conda-forge:
```
mamba install py2neo neotime
```
#### For jupyterlab 1.x or 2.x:
If you are using JupyterLab 1.x or 2.x then you will also need to install `nodejs` and the `jupyterlab-manager` extension. You can do this like so:
```bash
# installing nodejs
conda install -c conda-forge nodejs
# install jupyterlab-manager extension
jupyter labextension install @jupyter-widgets/jupyterlab-manager@2.0 --no-build
# if you have previously installed the manager you still to run jupyter lab build
jupyter lab build
```
### For Jupyter Notebook 5.2 and earlier
You may also need to manually enable the nbextension:
```bash
jupyter nbextension enable --py [--sys-prefix|--user|--system] ipycytoscape
```
## For a development installation:
**(requires npm)**
While not required, we recommend creating a conda environment to work in:
```bash
conda create -n ipycytoscape -c conda-forge jupyterlab nodejs
conda activate ipycytoscape
# clone repo
git clone https://github.com/cytoscape/ipycytoscape.git
cd ipycytoscape
```
### Install python package for development
This will `run npm install` and `npm run build`.
This command will also install the test suite and the [docs](https://ipycytoscape.readthedocs.io/en/latest/) locally:
```
pip install jupyter_packaging==0.7.9
pip install -e ".[test, docs]"
jupyter labextension develop . --overwrite
```
optionally install the pre-commit hooks with:
```bash
pre-commit install
```
Or for classic notebook, you can run:
```
jupyter nbextension install --sys-prefix --symlink --overwrite --py ipycytoscape
jupyter nbextension enable --sys-prefix --py ipycytoscape
```
Note that the `--symlink` flag doesn't work on Windows, so you will here have to run
the `install` command every time that you rebuild your extension. For certain installations
you might also need another flag instead of `--sys-prefix`, but we won't cover the meaning
of those flags here.
### How to see your changes
#### Typescript:
To continuously monitor the project for changes and automatically trigger a rebuild, start watching the ipycytoscape code:
```bash
npm run watch
```
And in a separate terminal start JupyterLab normally:
```bash
jupyter lab
```
once the webpack rebuild finishes refresh the JupyterLab page to have your changes take effect.
#### Python:
If you make a change to the python code then you need to restart the notebook kernel to have it take effect.
### How to run tests locally
Install necessary dependencies with pip:
```
pip install -e ".[test]"
```
Or with conda/mamba:
```
mamba -c conda-forge install networkx pandas nbval pytest
```
#### And to run it:
```
pytest
```
### How to build the docs
`cd docs`
Install dependencies:
`conda env update --file doc_environment.yml`
And build them:
`make html`
## Acknowledgements
The ipycytoscape project was started by [Mariana Meireles](https://github.com/marimeireles) at [QuantStack](https://quantstack.net). This initial development was funded as part of the [PLASMA](https://plasmabio.org) project, which is led by Claire Vandiedonck, Pierre Poulain, and Sandrine Caburet.
## License
We use a shared copyright model that enables all contributors to maintain the
copyright on their contributions.
This software is licensed under the BSD-3-Clause license. See the
[LICENSE](LICENSE) file for details.
Raw data
{
"_id": null,
"home_page": "https://github.com/cytoscape/ipycytoscape",
"name": "ipycytoscape",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "Jupyter,Widgets,IPython",
"author": "Mariana Meireles",
"author_email": "mariana.meireles@quantstack.net",
"download_url": "https://files.pythonhosted.org/packages/21/4b/dca529aa566ce225107580c6c8625c7dc5ecb1532f7d73259e2888d2187a/ipycytoscape-1.3.3.tar.gz",
"platform": "Linux",
"description": "# ipycytoscape\n\n[](https://github.com/cytoscape/ipycytoscape/actions/workflows/test.yml) [](https://ipycytoscape.readthedocs.io/en/latest/?badge=master) [](https://stackoverflow.com/questions/tagged/ipycytoscape) [](https://gitter.im/QuantStack/Lobby?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge)\n\nA widget enabling interactive graph visualization with [cytoscape.js](https://js.cytoscape.org/) in JupyterLab and the Jupyter notebook.\n\nTry it out using binder: [](https://mybinder.org/v2/gh/cytoscape/ipycytoscape/HEAD?filepath=examples) or install and try out the [examples](examples).\n\n\n\n#### Supports:\n\n* Conversion from NetworkX see [example1](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Test%20NetworkX%20methods.ipynb), [example2](https://github.com/cytoscape/ipycytoscape/blob/master/examples/NetworkX%20Example.ipynb)\n* Conversion from Pandas DataFrame see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/pandas.ipynb)\n* Conversion from neo4j see [example](https://github.com/cytoscape/ipycytoscape/blob/master/examples/Neo4j_Example.ipynb)\n\n## Installation\n\nWith `mamba`:\n\n```\nmamba install -c conda-forge ipycytoscape\n```\n\nWith `conda`:\n\n```\nconda install -c conda-forge ipycytoscape\n```\n\nWith `pip`:\n\n```bash\npip install ipycytoscape\n```\n\n### Pandas installation\n\nYou can install the Pandas dependencies for `ipycytoscape` with pip:\n\n```\npip install pandas\n```\n\nOr conda-forge:\n\n```\nmamba install pandas\n```\n\n### Neo4j installation\n\nYou can install the neo4j dependencies for `ipycytoscape` with pip:\n\n```\npip install -e \".[neo4j]\"\n```\n\nOr conda-forge:\n```\nmamba install py2neo neotime\n```\n\n#### For jupyterlab 1.x or 2.x:\n\nIf you are using JupyterLab 1.x or 2.x then you will also need to install `nodejs` and the `jupyterlab-manager` extension. You can do this like so:\n\n```bash\n# installing nodejs\nconda install -c conda-forge nodejs\n\n\n# install jupyterlab-manager extension\njupyter labextension install @jupyter-widgets/jupyterlab-manager@2.0 --no-build\n\n# if you have previously installed the manager you still to run jupyter lab build\njupyter lab build\n```\n\n### For Jupyter Notebook 5.2 and earlier\n\nYou may also need to manually enable the nbextension:\n```bash\njupyter nbextension enable --py [--sys-prefix|--user|--system] ipycytoscape\n```\n\n## For a development installation:\n**(requires npm)**\n\nWhile not required, we recommend creating a conda environment to work in:\n```bash\nconda create -n ipycytoscape -c conda-forge jupyterlab nodejs\nconda activate ipycytoscape\n\n# clone repo\ngit clone https://github.com/cytoscape/ipycytoscape.git\ncd ipycytoscape\n```\n\n### Install python package for development\n\nThis will `run npm install` and `npm run build`. \nThis command will also install the test suite and the [docs](https://ipycytoscape.readthedocs.io/en/latest/) locally:\n\n```\npip install jupyter_packaging==0.7.9\npip install -e \".[test, docs]\"\n\njupyter labextension develop . --overwrite\n```\n\noptionally install the pre-commit hooks with:\n\n```bash\npre-commit install\n```\n\n\nOr for classic notebook, you can run:\n\n```\njupyter nbextension install --sys-prefix --symlink --overwrite --py ipycytoscape\njupyter nbextension enable --sys-prefix --py ipycytoscape\n```\n\nNote that the `--symlink` flag doesn't work on Windows, so you will here have to run\nthe `install` command every time that you rebuild your extension. For certain installations\nyou might also need another flag instead of `--sys-prefix`, but we won't cover the meaning\nof those flags here.\n\n### How to see your changes\n\n#### Typescript: \nTo continuously monitor the project for changes and automatically trigger a rebuild, start watching the ipycytoscape code:\n```bash\nnpm run watch\n```\nAnd in a separate terminal start JupyterLab normally:\n```bash\njupyter lab\n```\nonce the webpack rebuild finishes refresh the JupyterLab page to have your changes take effect.\n\n#### Python:\nIf you make a change to the python code then you need to restart the notebook kernel to have it take effect.\n\n### How to run tests locally\n\nInstall necessary dependencies with pip:\n\n```\npip install -e \".[test]\"\n```\n\nOr with conda/mamba:\n\n```\nmamba -c conda-forge install networkx pandas nbval pytest\n```\n\n#### And to run it:\n\n```\npytest\n```\n\n### How to build the docs\n\n`cd docs`\n\nInstall dependencies:\n\n`conda env update --file doc_environment.yml`\n\nAnd build them: \n\n`make html`\n\n## Acknowledgements\n\nThe ipycytoscape project was started by [Mariana Meireles](https://github.com/marimeireles) at [QuantStack](https://quantstack.net). This initial development was funded as part of the [PLASMA](https://plasmabio.org) project, which is led by Claire Vandiedonck, Pierre Poulain, and Sandrine Caburet.\n\n## License\n\nWe use a shared copyright model that enables all contributors to maintain the\ncopyright on their contributions.\n\nThis software is licensed under the BSD-3-Clause license. See the\n[LICENSE](LICENSE) file for details.\n\n\n",
"bugtrack_url": null,
"license": "BSD",
"summary": "A Cytoscape widget for Jupyter",
"version": "1.3.3",
"split_keywords": [
"jupyter",
"widgets",
"ipython"
],
"urls": [
{
"comment_text": "",
"digests": {
"md5": "329dffcf27381fb4585f11a23f9aa98c",
"sha256": "4bc205724971f5f7a3fc2b09dfec20c357c4c6dfa2b4bd41e7c33c995c3f6906"
},
"downloads": -1,
"filename": "ipycytoscape-1.3.3-py2.py3-none-any.whl",
"has_sig": false,
"md5_digest": "329dffcf27381fb4585f11a23f9aa98c",
"packagetype": "bdist_wheel",
"python_version": "py2.py3",
"requires_python": null,
"size": 3595634,
"upload_time": "2022-04-04T22:26:36",
"upload_time_iso_8601": "2022-04-04T22:26:36.926571Z",
"url": "https://files.pythonhosted.org/packages/4c/0f/b66d63d4a5426c09005d3713b056e634e00e69788fdc88d1ffe40e5b7654/ipycytoscape-1.3.3-py2.py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "803c83a9c3663260bac095f4c6a27d80",
"sha256": "b6f3199df034f088e92d388e27e629f58ae2901b213cb9299e5b564272f9a2f8"
},
"downloads": -1,
"filename": "ipycytoscape-1.3.3.tar.gz",
"has_sig": false,
"md5_digest": "803c83a9c3663260bac095f4c6a27d80",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 3885550,
"upload_time": "2022-04-04T22:26:45",
"upload_time_iso_8601": "2022-04-04T22:26:45.943919Z",
"url": "https://files.pythonhosted.org/packages/21/4b/dca529aa566ce225107580c6c8625c7dc5ecb1532f7d73259e2888d2187a/ipycytoscape-1.3.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2022-04-04 22:26:45",
"github": true,
"gitlab": false,
"bitbucket": false,
"github_user": "cytoscape",
"github_project": "ipycytoscape",
"travis_ci": false,
"coveralls": true,
"github_actions": true,
"appveyor": true,
"lcname": "ipycytoscape"
}

