# ipyspeck
## ipyspeck Stats
<table>
<tr>
<td>Latest Release</td>
<td>
<a href="https://pypi.org/project/ipyspeck/"/>
<img src="https://badge.fury.io/py/ipyspeck.svg"/>
</td>
</tr>
<tr>
<td>PyPI Downloads</td>
<td>
<a href="https://pepy.tech/project/ipyspeck"/>
<img src="https://static.pepy.tech/badge/ipyspeck/month"/>
<img src="https://static.pepy.tech/badge/ipyspeck"/>
</td>
</tr>
</table>
## Speck
Speck is a molecule renderer with the goal of producing figures that are as attractive as they are practical. Express your molecule clearly _and_ with style.
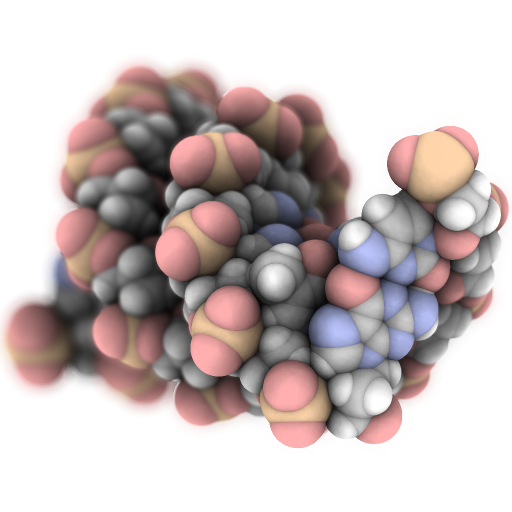
## ipypeck
Ipyspeck is a ipywidget wrapping speck to be used on a Jupyter notebook as a regular widget.
## Usage
The ipyspeck widget renders xyz molecules.
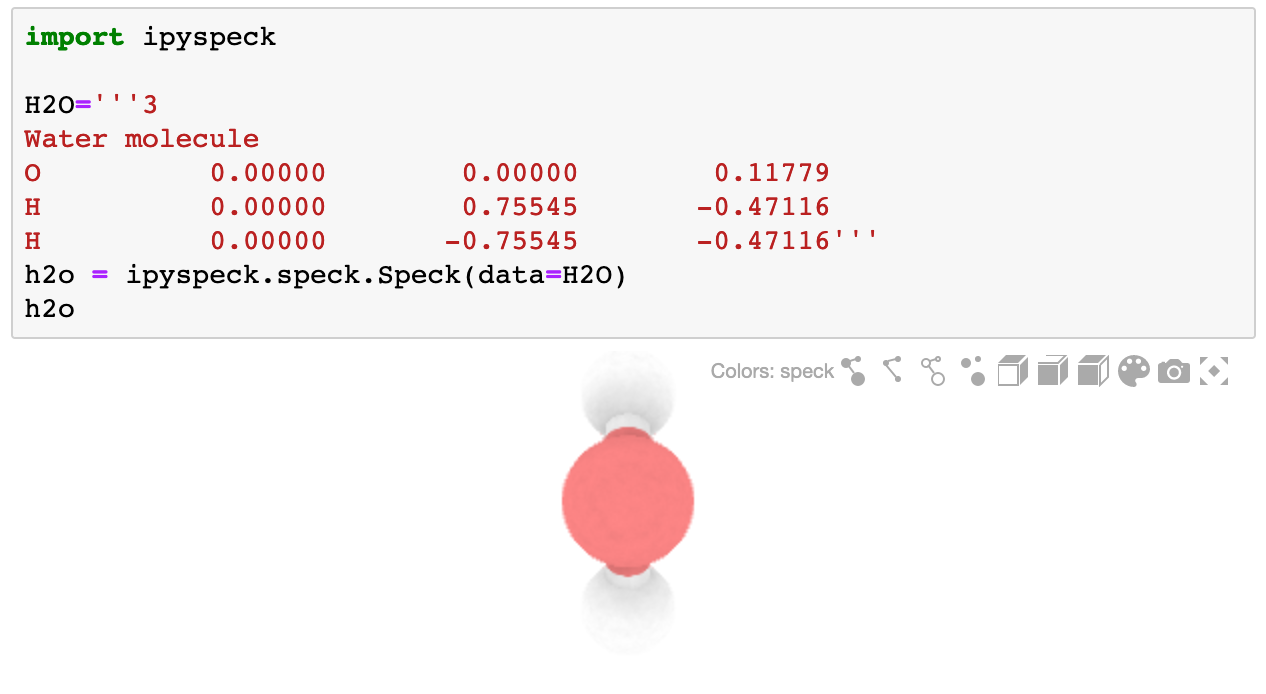
```bash
import ipyspeck
H2O='''3
Water molecule
O 0.00000 0.00000 0.11779
H 0.00000 0.75545 -0.47116
H 0.00000 -0.75545 -0.47116'''
h2o = ipyspeck.speck.Speck(data=H2O)
h2o
```
Ideally it should be used as part of a container widget (such as Box, VBox, Grid, ...)
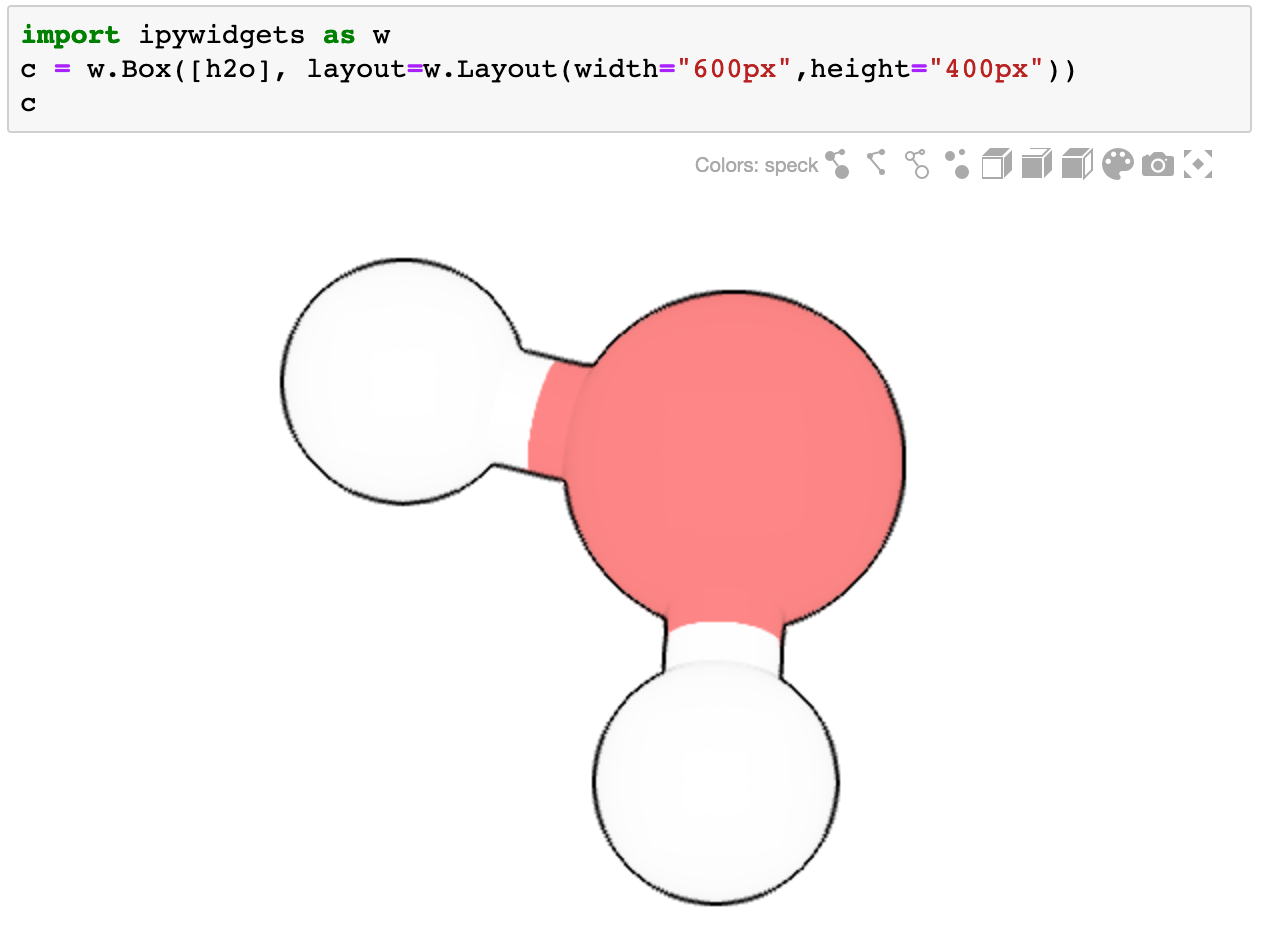
```bash
import ipywidgets as w
c = w.Box([h2o], layout=w.Layout(width="600px",height="400px"))
c
```
The visualization parameters can be modified
```bash
#Modify atoms size
h2o.atomScale = 0.3
#change bonds size
h2o.bondScale = 0.3
#highlight borders
h2o.outline = 0
```
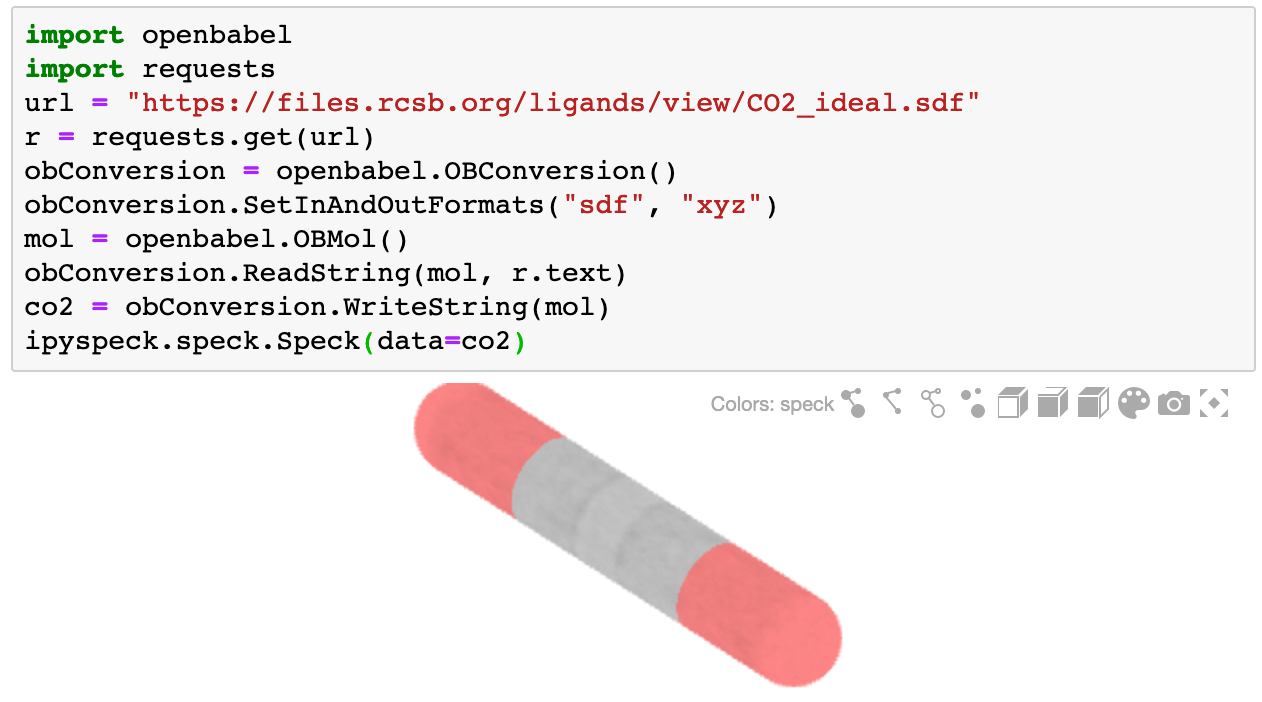
To render molecules on different formats openbabel can be used to translate them as xyz
```bash
import openbabel
import requests
url = "https://files.rcsb.org/ligands/view/CO2_ideal.sdf"
r = requests.get(url)
obConversion = openbabel.OBConversion()
obConversion.SetInAndOutFormats("sdf", "xyz")
mol = openbabel.OBMol()
obConversion.ReadString(mol, r.text)
co2 = obConversion.WriteString(mol)
ipyspeck.speck.Speck(data=co2)
```
## Installation
To install use pip:
$ pip install ipyspeck
$ jupyter nbextension enable --py --sys-prefix ipyspeck
To install for jupyterlab
$ jupyter labextension install ipyspeck
For a development installation (requires npm),
$ git clone https://github.com//denphi//speck.git
$ cd speck/widget/ipyspeck
$ pip install -e .
$ jupyter nbextension install --py --symlink --sys-prefix ipyspeck
$ jupyter nbextension enable --py --sys-prefix ipyspeck
$ jupyter labextension install js
When actively developing your extension, build Jupyter Lab with the command:
$ jupyter lab --watch
This takes a minute or so to get started, but then automatically rebuilds JupyterLab when your javascript changes.
Note on first `jupyter lab --watch`, you may need to touch a file to get Jupyter Lab to open.
## Gallery
<img src="https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/loop.gif" width=500px/>
<img src="https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img1.png" width=500px/>
<img src="https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img2.png" width=500px/>
<img src="https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img3.png" width=500px/>
<img src="https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img4.png" width=500px/>
Raw data
{
"_id": null,
"home_page": "https://github.com/denphi/speck/tree/master/widget/ipyspeck",
"name": "ipyspeck",
"maintainer": null,
"docs_url": null,
"requires_python": null,
"maintainer_email": null,
"keywords": "ipython, jupyter, widgets",
"author": "Daniel Mejia (Denphi)",
"author_email": "denphi@denphi.com",
"download_url": "https://files.pythonhosted.org/packages/6e/b7/9e2886e5a460d66dcec49c6a1247975e881ef7669303dc81dfb7ae58cced/ipyspeck-0.6.2.tar.gz",
"platform": null,
"description": "# ipyspeck\n\n## ipyspeck Stats\n\n<table>\n <tr>\n <td>Latest Release</td>\n <td>\n <a href=\"https://pypi.org/project/ipyspeck/\"/>\n <img src=\"https://badge.fury.io/py/ipyspeck.svg\"/>\n </td>\n </tr>\n <tr>\n <td>PyPI Downloads</td>\n <td>\n <a href=\"https://pepy.tech/project/ipyspeck\"/>\n <img src=\"https://static.pepy.tech/badge/ipyspeck/month\"/>\n <img src=\"https://static.pepy.tech/badge/ipyspeck\"/>\n </td>\n </tr>\n</table>\n\n## Speck\n\nSpeck is a molecule renderer with the goal of producing figures that are as attractive as they are practical. Express your molecule clearly _and_ with style.\n\n\n\n## ipypeck\n\nIpyspeck is a ipywidget wrapping speck to be used on a Jupyter notebook as a regular widget.\n\n## Usage\n\nThe ipyspeck widget renders xyz molecules.\n\n\n\n```bash\nimport ipyspeck\n\nH2O='''3\nWater molecule\nO 0.00000 0.00000 0.11779\nH 0.00000 0.75545 -0.47116\nH 0.00000 -0.75545 -0.47116'''\nh2o = ipyspeck.speck.Speck(data=H2O)\nh2o\n```\n\nIdeally it should be used as part of a container widget (such as Box, VBox, Grid, ...)\n\n\n\n\n```bash\n\nimport ipywidgets as w\nc = w.Box([h2o], layout=w.Layout(width=\"600px\",height=\"400px\"))\nc\n```\n\nThe visualization parameters can be modified\n```bash\n#Modify atoms size\nh2o.atomScale = 0.3\n#change bonds size\nh2o.bondScale = 0.3\n#highlight borders\nh2o.outline = 0\n```\n\nTo render molecules on different formats openbabel can be used to translate them as xyz\n\n```bash\nimport openbabel\nimport requests\nurl = \"https://files.rcsb.org/ligands/view/CO2_ideal.sdf\"\nr = requests.get(url)\nobConversion = openbabel.OBConversion()\nobConversion.SetInAndOutFormats(\"sdf\", \"xyz\")\nmol = openbabel.OBMol()\nobConversion.ReadString(mol, r.text)\nco2 = obConversion.WriteString(mol)\nipyspeck.speck.Speck(data=co2)\n```\n\n\n\n## Installation\n\nTo install use pip:\n\n $ pip install ipyspeck\n $ jupyter nbextension enable --py --sys-prefix ipyspeck\n\nTo install for jupyterlab\n\n $ jupyter labextension install ipyspeck\n\nFor a development installation (requires npm),\n\n $ git clone https://github.com//denphi//speck.git\n $ cd speck/widget/ipyspeck\n $ pip install -e .\n $ jupyter nbextension install --py --symlink --sys-prefix ipyspeck\n $ jupyter nbextension enable --py --sys-prefix ipyspeck\n $ jupyter labextension install js\n\nWhen actively developing your extension, build Jupyter Lab with the command:\n\n $ jupyter lab --watch\n\nThis takes a minute or so to get started, but then automatically rebuilds JupyterLab when your javascript changes.\n\nNote on first `jupyter lab --watch`, you may need to touch a file to get Jupyter Lab to open.\n\n## Gallery\n\n<img src=\"https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/loop.gif\" width=500px/>\n\n<img src=\"https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img1.png\" width=500px/>\n\n<img src=\"https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img2.png\" width=500px/>\n\n<img src=\"https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img3.png\" width=500px/>\n\n<img src=\"https://raw.githubusercontent.com/denphi/speck/master/widget/ipyspeck/img/img4.png\" width=500px/>\n",
"bugtrack_url": null,
"license": null,
"summary": "Speck Jupyter Widget",
"version": "0.6.2",
"project_urls": {
"Homepage": "https://github.com/denphi/speck/tree/master/widget/ipyspeck"
},
"split_keywords": [
"ipython",
" jupyter",
" widgets"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "dfa8fdb716521c0a2a3dd9470f07297152c0edc1bae3d4f8ce749f6b7b76f495",
"md5": "913e5c8764e5afc438404b762efebbbf",
"sha256": "a6eac3453b48bb3c6d91beadec7ac363a380bf1b4c0f055d754031ac2426ef8c"
},
"downloads": -1,
"filename": "ipyspeck-0.6.2-py2.py3-none-any.whl",
"has_sig": false,
"md5_digest": "913e5c8764e5afc438404b762efebbbf",
"packagetype": "bdist_wheel",
"python_version": "py2.py3",
"requires_python": null,
"size": 2380127,
"upload_time": "2024-05-16T20:44:30",
"upload_time_iso_8601": "2024-05-16T20:44:30.290982Z",
"url": "https://files.pythonhosted.org/packages/df/a8/fdb716521c0a2a3dd9470f07297152c0edc1bae3d4f8ce749f6b7b76f495/ipyspeck-0.6.2-py2.py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "6eb79e2886e5a460d66dcec49c6a1247975e881ef7669303dc81dfb7ae58cced",
"md5": "b3d4706ad8170a3707486fe3624aee02",
"sha256": "b28587ac294564f5855fbea0ebb077b33f194008a9126ca37e0f7a24fd8e5e58"
},
"downloads": -1,
"filename": "ipyspeck-0.6.2.tar.gz",
"has_sig": false,
"md5_digest": "b3d4706ad8170a3707486fe3624aee02",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 883592,
"upload_time": "2024-05-16T20:44:32",
"upload_time_iso_8601": "2024-05-16T20:44:32.472397Z",
"url": "https://files.pythonhosted.org/packages/6e/b7/9e2886e5a460d66dcec49c6a1247975e881ef7669303dc81dfb7ae58cced/ipyspeck-0.6.2.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-05-16 20:44:32",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "denphi",
"github_project": "speck",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "ipyspeck"
}
