# meshed
Object composition.
In particular: Link functions up into callable objects (e.g. pipelines, DAGs, etc.)
To install: `pip install meshed`
[Documentation](https://i2mint.github.io/meshed/)
Note: The initial focuus of `meshed` was on DAGs, a versatile and probably most known kind of composition of functions,
but `meshed` aims at capturing much more than that.
# Quick Start
```python
from meshed import DAG
def this(a, b=1):
return a + b
def that(x, b=1):
return x * b
def combine(this, that):
return (this, that)
dag = DAG((this, that, combine))
print(dag.synopsis_string())
```
x,b -> that_ -> that
a,b -> this_ -> this
this,that -> combine_ -> combine
But what does it do?
It's a callable, with a signature:
```python
from inspect import signature
signature(dag)
```
<Signature (x, a, b=1)>
And when you call it, it executes the dag from the root values you give it and
returns the leaf output values.
```python
dag(1, 2, 3) # (a+b,x*b) == (2+3,1*3) == (5, 3)
```
(5, 3)
```python
dag(1, 2) # (a+b,x*b) == (2+1,1*1) == (3, 1)
```
(3, 1)
You can see (and save image, or ascii art) the dag:
```python
dag.dot_digraph()
```
<img src="https://user-images.githubusercontent.com/1906276/127779463-ae75604b-0d69-4ac4-b206-80c2c5ae582b.png" width=200>
You can extend a dag
```python
dag2 = DAG([*dag, lambda this, a: this + a])
dag2.dot_digraph()
```
<img src="https://user-images.githubusercontent.com/1906276/127779748-70b47907-e51f-4e64-bc18-9545ee07e632.png" width=200>
You can get a sub-dag by specifying desired input(s) and outputs.
```python
dag2[['that', 'this'], 'combine'].dot_digraph()
```
<img src="https://user-images.githubusercontent.com/1906276/127779781-8aac40eb-ed52-4694-b50e-4af896cc30a2.png" width=150>
## Note on flexibility
The above DAG was created straight from the functions, using only the names of the
functions and their parameters to define how to hook the network up.
But if you didn't write those functions specifically for that purpose, or you want
to use someone else's functions, one would need to specify the relation between parameters, inputs and outputs.
For that purpose, functions can be adapted using the class FuncNode. The class allows you to essentially rename each of the parameters and also specify which output should be used as an argument for any other functions.
Let us consider the example below.
```python
def f(a, b):
return a + b
def g(a_plus_b, d):
return a_plus_b * d
```
Say we want the output of f to become the value of the parameter a_plus_b. We can do that by assigning the string 'a_plus_b' to the out parameter of a FuncNode representing the function f:
```python
f_node = FuncNode(func=f, out="a_plus_b")
```
We can now create a dag using our f_node instead of f:
```python
dag = DAG((f_node, g))
```
Our dag behaves as wanted:
```python
dag(a=1, b=2, d=3)
9
```
Now say we would also like for the value given to b to be also given to d. We can achieve that by binding d to b in the bind parameter of a FuncNode representing g:
```python
g_node = FuncNode(func=g, bind={"d": "b"})
```
The dag created with f_node and g_node has only two parameters, namely a and b:
```python
dag = DAG((f_node, g_node))
dag(a=1, b=2)
6
```
# Sub-DAGs
``dag[input_nodes:output_nodes]`` is the sub-dag made of intersection of all
descendants of ``input_nodes``
(inclusive) and ancestors of ``output_nodes`` (inclusive), where additionally,
when a func node is contained, it takes with it the input and output nodes
it needs.
```python
from meshed import DAG
def f(a): ...
def g(f): ...
def h(g): ...
def i(h): ...
dag = DAG([f, g, h, i])
dag.dot_digraph()
```
<img width="110" alt="image" src="https://user-images.githubusercontent.com/1906276/154749811-f9892ee6-617c-4fa6-9de9-1ebc509c04ae.png">
Get a subdag from ``g_`` (indicates the function here) to the end of ``dag``
```python
subdag = dag['g_',:]
subdag.dot_digraph()
```
<img width="100" alt="image" src="https://user-images.githubusercontent.com/1906276/154749842-c2320d1c-368d-4be8-ac57-9a77f1bb081d.png">
From the beginning to ``h_``
```python
dag[:, 'h_'].dot_digraph()
```
<img width="110" alt="image" src="https://user-images.githubusercontent.com/1906276/154750524-ece7f4b6-a3f3-46c6-a66d-7dc9b8ef254a.png">
From ``g_`` to ``h_`` (both inclusive)
```python
dag['g_', 'h_'].dot_digraph()
```
<img width="109" alt="image" src="https://user-images.githubusercontent.com/1906276/154749864-5a33aa13-0949-4aa7-945c-4d3fe7f07e7d.png">
Above we used function (node names) to specify what we wanted, but we can also
use names of input/output var-nodes. Do note the difference though.
The nodes you specify to get a sub-dag are INCLUSIVE, but when you
specify function nodes, you also get the input and output nodes of these
functions.
The ``dag['g_', 'h_']`` give us a sub-dag starting at ``f`` (the input node),
but when we ask ``dag['g', 'h_']`` instead, ``g`` being the output node of
function node ``g_``, we only get ``g -> h_ -> h``:
```python
dag['g', 'h'].dot_digraph()
```
<img width="88" alt="image" src="https://user-images.githubusercontent.com/1906276/154750753-737e2705-0ea3-4595-a93a-1567862a6edd.png">
If we wanted to include ``f`` we'd have to specify it:
```python
dag['f', 'h'].dot_digraph()
```
<img width="109" alt="image" src="https://user-images.githubusercontent.com/1906276/154749864-5a33aa13-0949-4aa7-945c-4d3fe7f07e7d.png">
Those were for simple pipelines, but let's now look at a more complex dag.
Note the definition: ``dag[input_nodes:output_nodes]`` is the sub-dag made of intersection of all
descendants of ``input_nodes``
(inclusive) and ancestors of ``output_nodes`` (inclusive), where additionally,
when a func node is contained, it takes with it the input and output nodes
it needs.
We'll let the following examples self-comment:
```python
from meshed import DAG
def f(u, v): ...
def g(f): ...
def h(f, w): ...
def i(g, h): ...
def j(h, x): ...
def k(i): ...
def l(i, j): ...
dag = DAG([f, g, h, i, j, k, l])
dag.dot_digraph()
```
<img width="248" alt="image" src="https://user-images.githubusercontent.com/1906276/154748574-a7026125-659f-465b-9bc3-14a1864d14b2.png">
```python
dag[['u', 'f'], 'h'].dot_digraph()
```
<img width="190" alt="image" src="https://user-images.githubusercontent.com/1906276/154748685-24e706ce-b68f-429a-b7b8-7bda62ccdf36.png">
```python
dag['u', 'h'].dot_digraph()
```
<img width="183" alt="image" src="https://user-images.githubusercontent.com/1906276/154748865-6e729094-976a-4af3-87f0-b6dd3900fb8c.png">
```python
dag[['u', 'f'], ['h', 'g']].dot_digraph()
```
<img width="199" alt="image" src="https://user-images.githubusercontent.com/1906276/154748905-4eaeccbe-6cca-4492-a7a2-48f7c9937b95.png">
```python
dag[['x', 'g'], 'k'].dot_digraph()
```
<img width="133" alt="image" src="https://user-images.githubusercontent.com/1906276/154748937-7a278b25-6f0f-467c-a977-89a175e15abb.png">
```python
dag[['x', 'g'], ['l', 'k']].dot_digraph()
```
<img width="216" alt="image" src="https://user-images.githubusercontent.com/1906276/154748958-135792a6-ce16-4561-9cbe-4662113a1022.png">
# Examples
## A train/test ML pipeline
Consider a simple train/test ML pipeline that looks like this.
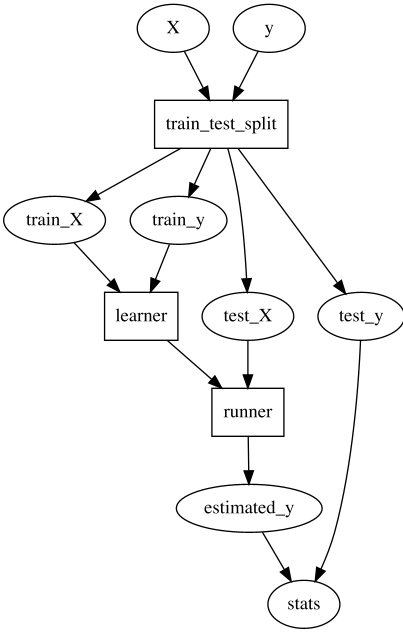
With this, we might decide we want to give the user control over how to do
`train_test_split` and `learner`, so we offer this interface to the user:
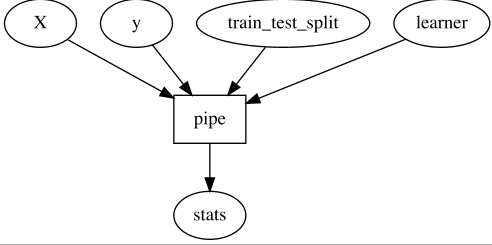
With that, the user can just bring its own `train_test_split` and `learner`
functions, and as long as it satisfied the
expected (and even better; declared and validatable) protocol, things will work.
In some situations we'd like to fix some of how `train_test_split` and
`learner` work, allowing the user to control only some aspects of them.
This function would look like this:
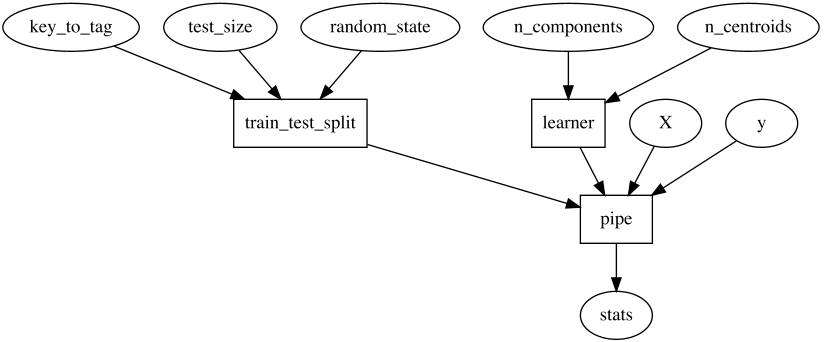
And inside, it does:
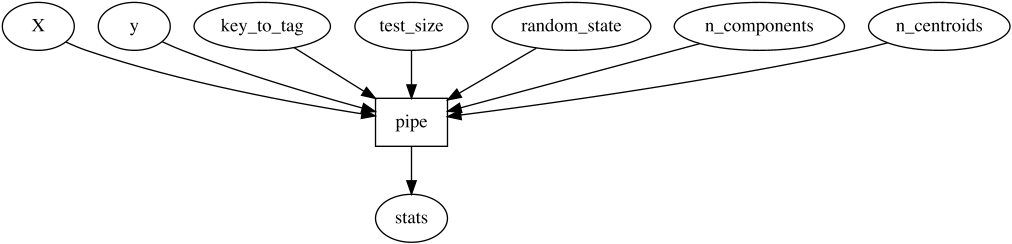
`meshed` allows us to easily manipulate such functional structures to
adapt them to our needs.
# itools module
Tools that enable operations on graphs where graphs are represented by an adjacency Mapping.
Again.
Graphs: You know them. Networks.
Nodes and edges, and the ecosystem descriptive or transformative functions surrounding these.
Few languages have builtin support for the graph data structure, but all have their libraries to compensate.
The one you're looking at focuses on the representation of a graph as `Mapping` encoding
its [adjacency list](https://en.wikipedia.org/wiki/Adjacency_list).
That is, a dictionary-like interface that specifies the graph by specifying for each node
what nodes it's adjacent to:
```python
assert graph[source_node] == set_of_nodes_that_source_node_has_edges_to
```
We emphasize that there is no specific graph instance that you need to squeeze your graph into to
be able to use the functions of `meshed`. Suffices that your graph's structure is expressed by
that dict-like interface
-- which grown-ups call `Mapping` (see the `collections.abc` or `typing` standard libs for more information).
You'll find a lot of `Mapping`s around pythons.
And if the object you want to work with doesn't have that interface,
you can easily create one using one of the many tools of `py2store` meant exactly for that purpose.
# Examples
```pydocstring
>>> from meshed.itools import edges, nodes, isolated_nodes
>>> graph = dict(a='c', b='ce', c='abde', d='c', e=['c', 'b'], f={})
>>> sorted(edges(graph))
[('a', 'c'), ('b', 'c'), ('b', 'e'), ('c', 'a'), ('c', 'b'), ('c', 'd'), ('c', 'e'), ('d', 'c'), ('e', 'b'), ('e', 'c')]
>>> sorted(nodes(graph))
['a', 'b', 'c', 'd', 'e', 'f']
>>> set(isolated_nodes(graph))
{'f'}
>>>
>>> from meshed.makers import edge_reversed_graph
>>> g = dict(a='c', b='cd', c='abd', e='')
>>> assert edge_reversed_graph(g) == {'c': ['a', 'b'], 'd': ['b', 'c'], 'a': ['c'], 'b': ['c'], 'e': []}
>>> reverse_g_with_sets = edge_reversed_graph(g, set, set.add)
>>> assert reverse_g_with_sets == {'c': {'a', 'b'}, 'd': {'b', 'c'}, 'a': {'c'}, 'b': {'c'}, 'e': set([])}
```
Raw data
{
"_id": null,
"home_page": "https://github.com/i2mint/meshed",
"name": "meshed",
"maintainer": null,
"docs_url": null,
"requires_python": null,
"maintainer_email": null,
"keywords": "dag, graph, network",
"author": null,
"author_email": null,
"download_url": "https://files.pythonhosted.org/packages/32/34/a0914dc1b0c84eaa2bd8f189dce7758128b471c11af45a6ea3d0e5536d0e/meshed-0.1.159.tar.gz",
"platform": "any",
"description": "# meshed\n\nObject composition. \nIn particular: Link functions up into callable objects (e.g. pipelines, DAGs, etc.)\n\nTo install: `pip install meshed`\n\n[Documentation](https://i2mint.github.io/meshed/)\n\nNote: The initial focuus of `meshed` was on DAGs, a versatile and probably most known kind of composition of functions, \nbut `meshed` aims at capturing much more than that. \n\n\n# Quick Start\n\n```python\nfrom meshed import DAG\n\ndef this(a, b=1):\n return a + b\ndef that(x, b=1):\n return x * b\ndef combine(this, that):\n return (this, that)\n\ndag = DAG((this, that, combine))\nprint(dag.synopsis_string())\n```\n\n x,b -> that_ -> that\n a,b -> this_ -> this\n this,that -> combine_ -> combine\n\n\nBut what does it do?\n\nIt's a callable, with a signature:\n\n```python\nfrom inspect import signature\nsignature(dag)\n```\n\n <Signature (x, a, b=1)>\n\nAnd when you call it, it executes the dag from the root values you give it and\nreturns the leaf output values.\n\n```python\ndag(1, 2, 3) # (a+b,x*b) == (2+3,1*3) == (5, 3)\n```\n (5, 3)\n\n```python\ndag(1, 2) # (a+b,x*b) == (2+1,1*1) == (3, 1)\n```\n (3, 1)\n\n\nYou can see (and save image, or ascii art) the dag:\n\n```python\ndag.dot_digraph()\n```\n\n<img src=\"https://user-images.githubusercontent.com/1906276/127779463-ae75604b-0d69-4ac4-b206-80c2c5ae582b.png\" width=200>\n\n\nYou can extend a dag\n\n```python\ndag2 = DAG([*dag, lambda this, a: this + a])\ndag2.dot_digraph()\n```\n\n<img src=\"https://user-images.githubusercontent.com/1906276/127779748-70b47907-e51f-4e64-bc18-9545ee07e632.png\" width=200>\n\nYou can get a sub-dag by specifying desired input(s) and outputs.\n\n```python\ndag2[['that', 'this'], 'combine'].dot_digraph()\n```\n\n<img src=\"https://user-images.githubusercontent.com/1906276/127779781-8aac40eb-ed52-4694-b50e-4af896cc30a2.png\" width=150>\n\n\n\n## Note on flexibility\n\nThe above DAG was created straight from the functions, using only the names of the\nfunctions and their parameters to define how to hook the network up.\n\nBut if you didn't write those functions specifically for that purpose, or you want\nto use someone else's functions, one would need to specify the relation between parameters, inputs and outputs.\n\nFor that purpose, functions can be adapted using the class FuncNode. The class allows you to essentially rename each of the parameters and also specify which output should be used as an argument for any other functions.\n\nLet us consider the example below.\n\n```python\ndef f(a, b):\n return a + b\n\ndef g(a_plus_b, d):\n return a_plus_b * d\n```\n\nSay we want the output of f to become the value of the parameter a_plus_b. We can do that by assigning the string 'a_plus_b' to the out parameter of a FuncNode representing the function f:\n\n```python\nf_node = FuncNode(func=f, out=\"a_plus_b\")\n```\n\nWe can now create a dag using our f_node instead of f:\n\n```python\ndag = DAG((f_node, g))\n```\n\nOur dag behaves as wanted:\n\n```python\ndag(a=1, b=2, d=3)\n9\n```\n\nNow say we would also like for the value given to b to be also given to d. We can achieve that by binding d to b in the bind parameter of a FuncNode representing g:\n\n```python\ng_node = FuncNode(func=g, bind={\"d\": \"b\"})\n```\n\nThe dag created with f_node and g_node has only two parameters, namely a and b:\n\n```python\ndag = DAG((f_node, g_node))\ndag(a=1, b=2)\n6\n```\n\n\n\n\n# Sub-DAGs\n\n\n``dag[input_nodes:output_nodes]`` is the sub-dag made of intersection of all\ndescendants of ``input_nodes``\n(inclusive) and ancestors of ``output_nodes`` (inclusive), where additionally,\nwhen a func node is contained, it takes with it the input and output nodes\nit needs.\n\n\n```python\nfrom meshed import DAG\n\ndef f(a): ...\ndef g(f): ...\ndef h(g): ...\ndef i(h): ...\ndag = DAG([f, g, h, i])\n\ndag.dot_digraph()\n```\n\n<img width=\"110\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154749811-f9892ee6-617c-4fa6-9de9-1ebc509c04ae.png\">\n\n\nGet a subdag from ``g_`` (indicates the function here) to the end of ``dag``\n\n```python\nsubdag = dag['g_',:]\nsubdag.dot_digraph()\n```\n\n<img width=\"100\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154749842-c2320d1c-368d-4be8-ac57-9a77f1bb081d.png\">\n\nFrom the beginning to ``h_``\n\n```python\ndag[:, 'h_'].dot_digraph()\n```\n\n<img width=\"110\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154750524-ece7f4b6-a3f3-46c6-a66d-7dc9b8ef254a.png\">\n\n\n\nFrom ``g_`` to ``h_`` (both inclusive)\n\n```python\ndag['g_', 'h_'].dot_digraph()\n```\n\n<img width=\"109\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154749864-5a33aa13-0949-4aa7-945c-4d3fe7f07e7d.png\">\n\n\nAbove we used function (node names) to specify what we wanted, but we can also\nuse names of input/output var-nodes. Do note the difference though.\nThe nodes you specify to get a sub-dag are INCLUSIVE, but when you\nspecify function nodes, you also get the input and output nodes of these\nfunctions.\n\nThe ``dag['g_', 'h_']`` give us a sub-dag starting at ``f`` (the input node),\nbut when we ask ``dag['g', 'h_']`` instead, ``g`` being the output node of\nfunction node ``g_``, we only get ``g -> h_ -> h``:\n\n```python\ndag['g', 'h'].dot_digraph()\n```\n\n<img width=\"88\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154750753-737e2705-0ea3-4595-a93a-1567862a6edd.png\">\n\n\nIf we wanted to include ``f`` we'd have to specify it:\n\n\n```python\ndag['f', 'h'].dot_digraph()\n```\n\n<img width=\"109\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154749864-5a33aa13-0949-4aa7-945c-4d3fe7f07e7d.png\">\n\n\nThose were for simple pipelines, but let's now look at a more complex dag.\n\nNote the definition: ``dag[input_nodes:output_nodes]`` is the sub-dag made of intersection of all \ndescendants of ``input_nodes``\n(inclusive) and ancestors of ``output_nodes`` (inclusive), where additionally,\nwhen a func node is contained, it takes with it the input and output nodes\nit needs.\n\nWe'll let the following examples self-comment:\n\n```python\nfrom meshed import DAG\n\n\ndef f(u, v): ...\n\ndef g(f): ...\n\ndef h(f, w): ...\n\ndef i(g, h): ...\n\ndef j(h, x): ...\n\ndef k(i): ...\n\ndef l(i, j): ...\n\ndag = DAG([f, g, h, i, j, k, l])\n\ndag.dot_digraph()\n```\n\n<img width=\"248\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154748574-a7026125-659f-465b-9bc3-14a1864d14b2.png\">\n\n```python\ndag[['u', 'f'], 'h'].dot_digraph()\n```\n\n<img width=\"190\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154748685-24e706ce-b68f-429a-b7b8-7bda62ccdf36.png\">\n\n\n```python\ndag['u', 'h'].dot_digraph()\n```\n\n<img width=\"183\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154748865-6e729094-976a-4af3-87f0-b6dd3900fb8c.png\">\n\n\n```python\ndag[['u', 'f'], ['h', 'g']].dot_digraph()\n```\n\n<img width=\"199\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154748905-4eaeccbe-6cca-4492-a7a2-48f7c9937b95.png\">\n\n\n```python\ndag[['x', 'g'], 'k'].dot_digraph()\n```\n\n<img width=\"133\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154748937-7a278b25-6f0f-467c-a977-89a175e15abb.png\">\n\n```python\ndag[['x', 'g'], ['l', 'k']].dot_digraph()\n```\n\n<img width=\"216\" alt=\"image\" src=\"https://user-images.githubusercontent.com/1906276/154748958-135792a6-ce16-4561-9cbe-4662113a1022.png\">\n\n\n\n# Examples\n\n## A train/test ML pipeline\n\nConsider a simple train/test ML pipeline that looks like this.\n\n\n\nWith this, we might decide we want to give the user control over how to do \n`train_test_split` and `learner`, so we offer this interface to the user:\n\n\n\nWith that, the user can just bring its own `train_test_split` and `learner` \nfunctions, and as long as it satisfied the \nexpected (and even better; declared and validatable) protocol, things will work. \n\nIn some situations we'd like to fix some of how `train_test_split` and \n`learner` work, allowing the user to control only some aspects of them. \nThis function would look like this:\n\n\n\nAnd inside, it does:\n\n\n\n`meshed` allows us to easily manipulate such functional structures to \nadapt them to our needs.\n\n\n# itools module\nTools that enable operations on graphs where graphs are represented by an adjacency Mapping.\n\nAgain. \n\nGraphs: You know them. Networks. \nNodes and edges, and the ecosystem descriptive or transformative functions surrounding these.\nFew languages have builtin support for the graph data structure, but all have their libraries to compensate.\n\nThe one you're looking at focuses on the representation of a graph as `Mapping` encoding \nits [adjacency list](https://en.wikipedia.org/wiki/Adjacency_list). \nThat is, a dictionary-like interface that specifies the graph by specifying for each node\nwhat nodes it's adjacent to:\n\n```python\nassert graph[source_node] == set_of_nodes_that_source_node_has_edges_to\n```\n\nWe emphasize that there is no specific graph instance that you need to squeeze your graph into to\nbe able to use the functions of `meshed`. Suffices that your graph's structure is expressed by \nthat dict-like interface \n-- which grown-ups call `Mapping` (see the `collections.abc` or `typing` standard libs for more information).\n\nYou'll find a lot of `Mapping`s around pythons. \nAnd if the object you want to work with doesn't have that interface, \nyou can easily create one using one of the many tools of `py2store` meant exactly for that purpose.\n\n\n# Examples\n\n```pydocstring\n>>> from meshed.itools import edges, nodes, isolated_nodes\n>>> graph = dict(a='c', b='ce', c='abde', d='c', e=['c', 'b'], f={})\n>>> sorted(edges(graph))\n[('a', 'c'), ('b', 'c'), ('b', 'e'), ('c', 'a'), ('c', 'b'), ('c', 'd'), ('c', 'e'), ('d', 'c'), ('e', 'b'), ('e', 'c')]\n>>> sorted(nodes(graph))\n['a', 'b', 'c', 'd', 'e', 'f']\n>>> set(isolated_nodes(graph))\n{'f'}\n>>>\n>>> from meshed.makers import edge_reversed_graph\n>>> g = dict(a='c', b='cd', c='abd', e='')\n>>> assert edge_reversed_graph(g) == {'c': ['a', 'b'], 'd': ['b', 'c'], 'a': ['c'], 'b': ['c'], 'e': []}\n>>> reverse_g_with_sets = edge_reversed_graph(g, set, set.add)\n>>> assert reverse_g_with_sets == {'c': {'a', 'b'}, 'd': {'b', 'c'}, 'a': {'c'}, 'b': {'c'}, 'e': set([])}\n```\n",
"bugtrack_url": null,
"license": "mit",
"summary": "Link functions up into callable objects",
"version": "0.1.159",
"project_urls": {
"Homepage": "https://github.com/i2mint/meshed"
},
"split_keywords": [
"dag",
" graph",
" network"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "8cce4687d9c07a8e5c990df977b5644086d4709bf29c0eef3473fddc0dcecccb",
"md5": "184830868f651b31d988c83430a58b23",
"sha256": "6ba4fdedd8f81ec9b7868cfcc1e0f313937b291097edda074a0ea3734ea368a4"
},
"downloads": -1,
"filename": "meshed-0.1.159-py3-none-any.whl",
"has_sig": false,
"md5_digest": "184830868f651b31d988c83430a58b23",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": null,
"size": 132920,
"upload_time": "2024-07-18T08:48:52",
"upload_time_iso_8601": "2024-07-18T08:48:52.858461Z",
"url": "https://files.pythonhosted.org/packages/8c/ce/4687d9c07a8e5c990df977b5644086d4709bf29c0eef3473fddc0dcecccb/meshed-0.1.159-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "3234a0914dc1b0c84eaa2bd8f189dce7758128b471c11af45a6ea3d0e5536d0e",
"md5": "d6f2d0a67d6efe13057bb7e1b2e053d5",
"sha256": "8198ef32cc9d95c9b9513cc797fff00b8a74e54623837aa504887b2b5a6fb185"
},
"downloads": -1,
"filename": "meshed-0.1.159.tar.gz",
"has_sig": false,
"md5_digest": "d6f2d0a67d6efe13057bb7e1b2e053d5",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 122006,
"upload_time": "2024-07-18T08:48:55",
"upload_time_iso_8601": "2024-07-18T08:48:55.666538Z",
"url": "https://files.pythonhosted.org/packages/32/34/a0914dc1b0c84eaa2bd8f189dce7758128b471c11af45a6ea3d0e5536d0e/meshed-0.1.159.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-07-18 08:48:55",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "i2mint",
"github_project": "meshed",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "meshed"
}
