| Name | mlconfgen JSON |
| Version |
0.2.2
 JSON
JSON |
| download |
| home_page | None |
| Summary | Shape-constrained molecule generation via Equivariant Diffusion and GCN |
| upload_time | 2025-08-21 08:51:28 |
| maintainer | None |
| docs_url | None |
| author | None |
| requires_python | >=3.10 |
| license | None |
| keywords |
rdkit
chemistry
diffusion
conformers
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
# ML Conformer Generator
<img src="https://raw.githubusercontent.com/Membrizard/ml_conformer_generator/main/assets/logo/mlconfgen_logo.png" width="200" style="display: block; margin: 0 10%;">
**ML Conformer Generator**
is a tool for shape-constrained molecule generation using an Equivariant Diffusion Model (EDM)
and a Graph Convolutional Network (GCN). It is designed to generate 3D molecular conformations
that are both chemically valid and spatially similar to a reference shape.
## Supported features
* **Shape-guided molecular generation**
Generate novel molecules that conform to arbitrary 3D shapes—such as protein binding pockets or custom-defined spatial regions.
* **Reference-based conformer similarity**
Create molecules conformations of which closely resemble a reference structure, supporting scaffold-hopping and ligand-based design workflows.
* **Fragment-based inpainting**
Fix specific substructures or fragments within a molecule and complete or grow the rest in a geometrically consistent manner.
---
## Installation
1. Install the package:
`pip install mlconfgen`
2. Load the weights from Huggingface
> https://huggingface.co/Membrizard/ml_conformer_generator
`edm_moi_chembl_15_39.pt`
`adj_mat_seer_chembl_15_39.pt`
---
## 🐍 Python API
See interactive examples: `./python_api_demo.ipynb`
```python
from rdkit import Chem
from mlconfgen import MLConformerGenerator, evaluate_samples
model = MLConformerGenerator(
edm_weights="./edm_moi_chembl_15_39.pt",
adj_mat_seer_weights="./adj_mat_seer_chembl_15_39.pt",
diffusion_steps=100,
)
reference = Chem.MolFromMolFile('./assets/demo_files/ceyyag.mol')
samples = model.generate_conformers(reference_conformer=reference, n_samples=20, variance=2)
aligned_reference, std_samples = evaluate_samples(reference, samples)
```
---
## 🚀 Overview
This solution employs:
- **Equivariant Diffusion Model (EDM) [[1]](https://doi.org/10.48550/arXiv.2203.17003)**: For generating atom coordinates and types under a shape constraint.
- **Graph Convolutional Network (GCN) [[2]](https://doi.org/10.1039/D3DD00178D)**: For predicting atom adjacency matrices.
- **Deterministic Standardization Pipeline**: For refining and validating generated molecules.
---
## 🧠 Model Training
- Trained on **1.6 million** compounds from the **ChEMBL** database.
- Filtered to molecules with **15–39 heavy atoms**.
- Supported elements: `H, C, N, O, F, P, S, Cl, Br`.
---
## 🧪 Standardization Pipeline
The generated molecules are post-processed through the following steps:
- Largest Fragment picker
- Valence check
- Kekulization
- RDKit sanitization
- Constrained Geometry optimization via **MMFF94** Molecular Dynamics
---
## 📏 Evaluation Pipeline
Aligns and Evaluates shape similarity between generated molecules and a reference using
**Shape Tanimoto Similarity [[3]](https://doi.org/10.1007/978-94-017-1120-3_5 )** via Gaussian Molecular Volume overlap.
> Hydrogens are ignored in both reference and generated samples for this metric.
---
## 📊 Performance (100 Denoising Steps)
*Tested on 100,000 samples using 1,000 CCDC Virtual Screening [[4]](https://www.ccdc.cam.ac.uk/support-and-resources/downloads/) reference compounds.*
### General Overview
- ⏱ **Avg time to generate 50 valid samples**: 11.46 sec (NVIDIA H100)
- ⚡️ **Generation speed**: 4.18 valid molecules/sec
- 💾 **GPU memory (per generation thread)**: Up to 14.0 GB (`float16` 39 atoms 100 samples)
- 📐 **Avg Shape Tanimoto Similarity**: 53.32%
- 🎯 **Max Shape Tanimoto Similarity**: 99.69%
- 🔬 **Avg Chemical Tanimoto Similarity (2-hop 2048-bit Morgan Fingerprints)**: 10.87%
- 🧬 **% Chemically novel (vs. training set)**: 99.84%
- ✔️ **% Valid molecules (post-standardization)**: 48%
- 🔁 **% Unique molecules in generated set**: 99.94%
- ⚡ **Average Strain (MMFF94)**: 2.36 kcal / mol
- 📎 **Fréchet Fingerprint Distance (2-hop 2048-bit Morgan Fingerprints)**:
- To ChEMBL: 4.13
- To PubChem: 2.64
- To ZINC (250k): 4.95
### PoseBusters [[5]](https://doi.org/10.1039/D3SC04185A) validity check results:
**Overall stats**:
- PB-valid molecules: **91.33 %**
**Detailed Problems**:
- position: 0.01 %
- mol_pred_loaded: 0.0 %
- sanitization: 0.01 %
- inchi_convertible: 0.01 %
- all_atoms_connected: 0.0 %
- bond_lengths: 0.24 %
- bond_angles: 0.70 %
- internal_steric_clash: 2.31 %
- aromatic_ring_flatness: 3.34 %
- non-aromatic_ring_non-flatness: 0.27 %
### Synthesizability of the generated compounds
#### SA Score [[6]](https://doi.org/10.1186/1758-2946-1-8)
*1 (easy to make) - 10 (very difficult to make)*
**Average SA Score**: **3.18**
<img src="https://raw.githubusercontent.com/Membrizard/ml_conformer_generator/main/assets/benchmarks/sa_score_dist.png" width="300">
---
## Generation Examples
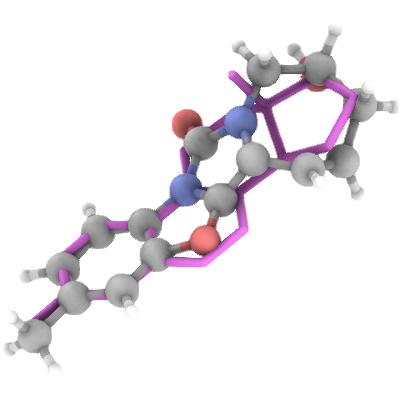
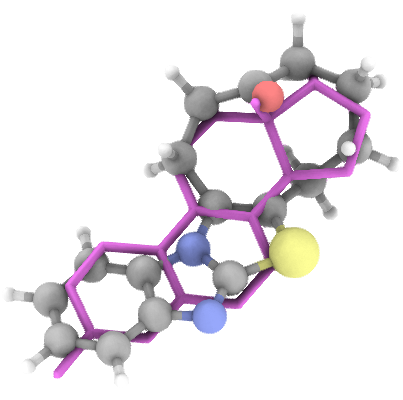
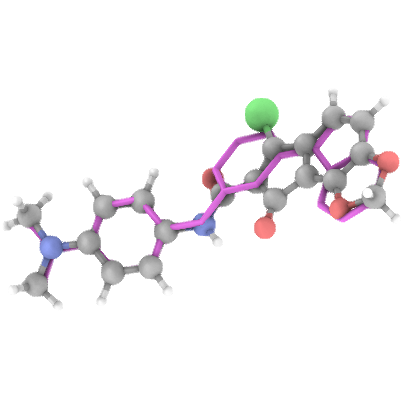
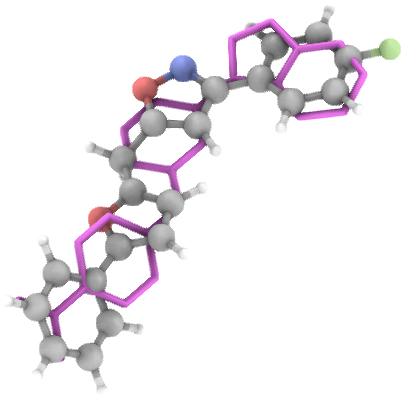
---
## 💾 Access & Licensing
The **Python package and inference code are available on GitHub** under Apache 2.0 License
> https://github.com/Membrizard/ml_conformer_generator
The trained model **Weights** are available at
> https://huggingface.co/Membrizard/ml_conformer_generator
And are licensed under CC BY-NC-ND 4.0
The usage of the trained weights for any profit-generating activity is restricted.
For commercial licensing and inference-as-a-service, contact:
[Denis Sapegin](https://github.com/Membrizard)
---
## ONNX Inference:
For torch Free inference an ONNX version of the model is present.
Weights of the model in ONNX format are available at:
> https://huggingface.co/Membrizard/ml_conformer_generator
`egnn_chembl_15_39.onnx`
`adj_mat_seer_chembl_15_39.onnx`
```python
from mlconfgen import MLConformerGeneratorONNX
from rdkit import Chem
model = MLConformerGeneratorONNX(
egnn_onnx="./egnn_chembl_15_39.onnx",
adj_mat_seer_onnx="./adj_mat_seer_chembl_15_39.onnx",
diffusion_steps=100,
)
reference = Chem.MolFromMolFile('./assets/demo_files/yibfeu.mol')
samples = model.generate_conformers(reference_conformer=reference, n_samples=20, variance=2)
```
Install ONNX GPU runtime (if needed):
`pip install onnxruntime-gpu`
---
## Export to ONNX
An option to compile the model to ONNX is provided
requires `onnxscript==0.2.2`
`pip install onnxscript`
```python
from mlconfgen import MLConformerGenerator
from onnx_export import export_to_onnx
model = MLConformerGenerator()
export_to_onnx(model)
```
This compiles and saves the ONNX files to: `./`
## Streamlit App
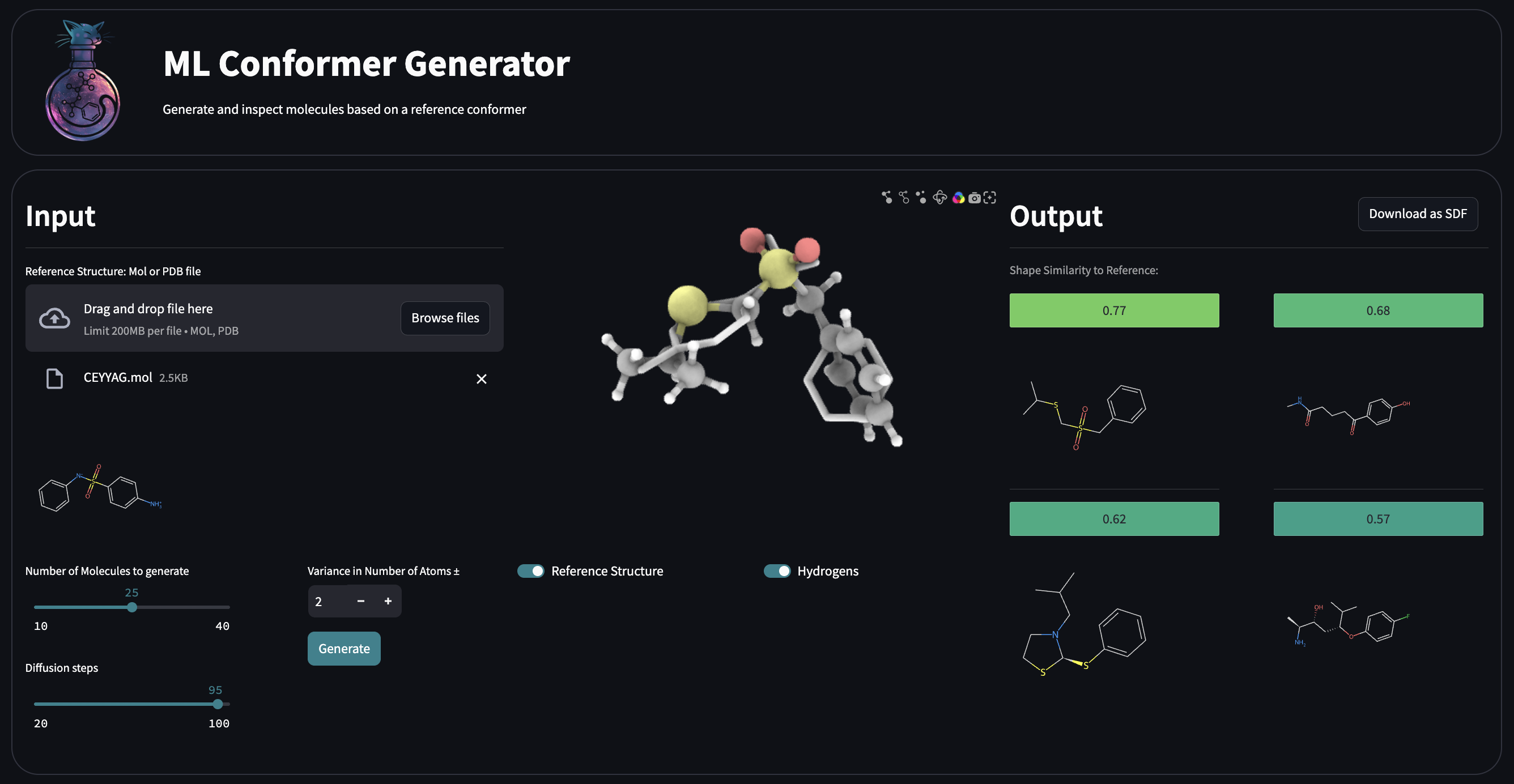
### Running
- Move the trained PyTorch weights into `./streamlit_app`
`./streamlit_app/edm_moi_chembl_15_39.pt`
`./streamlit_app/adj_mat_seer_chembl_15_39.pt`
- Install the dependencies `pip install -r ./streamlit_app/requirements.txt`
- Bring the app UI up:
```commandline
cd ./streamlit_app
streamlit run app.py
```
### Streamlit App Development
1. To enable development mode for the 3D viewer (`stspeck`), set `_RELEASE = False` in `./streamlit/stspeck/__init__.py`.
2. Navigate to the 3D viewer frontend and start the development server:
```commandline
cd ./frontend/speck/frontend
npm run start
```
This will launch the dev server at `http://localhost:3001`
3. In a separate terminal, run the Streamlit app from the root frontend directory:
```commandline
cd ./streamlit_app
streamlit run app.py
```
4. To build the production version of the 3D viewer, run:
```commandline
cd ./streamlit_app/stspeck/frontend
npm run build
```
Raw data
{
"_id": null,
"home_page": null,
"name": "mlconfgen",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": "Denis Sapegin <dasapegin@gmail.com>, Azamat Gafurov <azamat.gafurov@gmail.com>",
"keywords": "rdkit, chemistry, diffusion, conformers",
"author": null,
"author_email": "Denis Sapegin <dasapegin@gmail.com>, Fedor Bakharev <fbakharev@gmail.com>, Azamat Gafurov <azamat.gafurov@gmail.com>",
"download_url": "https://files.pythonhosted.org/packages/39/e4/baa776d8ba3eef4df7f7165474493be137133fbc14ff0abf1f1e494541e9/mlconfgen-0.2.2.tar.gz",
"platform": null,
"description": "# ML Conformer Generator\n\n<img src=\"https://raw.githubusercontent.com/Membrizard/ml_conformer_generator/main/assets/logo/mlconfgen_logo.png\" width=\"200\" style=\"display: block; margin: 0 10%;\">\n\n**ML Conformer Generator** \nis a tool for shape-constrained molecule generation using an Equivariant Diffusion Model (EDM)\nand a Graph Convolutional Network (GCN). It is designed to generate 3D molecular conformations\nthat are both chemically valid and spatially similar to a reference shape.\n\n## Supported features\n\n* **Shape-guided molecular generation**\n\n Generate novel molecules that conform to arbitrary 3D shapes\u2014such as protein binding pockets or custom-defined spatial regions.\n\n\n* **Reference-based conformer similarity**\n\n Create molecules conformations of which closely resemble a reference structure, supporting scaffold-hopping and ligand-based design workflows.\n\n\n* **Fragment-based inpainting**\n\n Fix specific substructures or fragments within a molecule and complete or grow the rest in a geometrically consistent manner.\n\n\n---\n## Installation\n\n1. Install the package:\n\n`pip install mlconfgen`\n\n2. Load the weights from Huggingface\n> https://huggingface.co/Membrizard/ml_conformer_generator\n\n`edm_moi_chembl_15_39.pt`\n\n`adj_mat_seer_chembl_15_39.pt`\n\n---\n\n## \ud83d\udc0d Python API\n\nSee interactive examples: `./python_api_demo.ipynb`\n\n```python\nfrom rdkit import Chem\nfrom mlconfgen import MLConformerGenerator, evaluate_samples\n\nmodel = MLConformerGenerator(\n edm_weights=\"./edm_moi_chembl_15_39.pt\",\n adj_mat_seer_weights=\"./adj_mat_seer_chembl_15_39.pt\",\n diffusion_steps=100,\n )\n\nreference = Chem.MolFromMolFile('./assets/demo_files/ceyyag.mol')\n\nsamples = model.generate_conformers(reference_conformer=reference, n_samples=20, variance=2)\n\naligned_reference, std_samples = evaluate_samples(reference, samples)\n```\n---\n\n## \ud83d\ude80 Overview\n\nThis solution employs:\n\n- **Equivariant Diffusion Model (EDM) [[1]](https://doi.org/10.48550/arXiv.2203.17003)**: For generating atom coordinates and types under a shape constraint.\n- **Graph Convolutional Network (GCN) [[2]](https://doi.org/10.1039/D3DD00178D)**: For predicting atom adjacency matrices.\n- **Deterministic Standardization Pipeline**: For refining and validating generated molecules.\n\n---\n\n## \ud83e\udde0 Model Training\n\n- Trained on **1.6 million** compounds from the **ChEMBL** database.\n- Filtered to molecules with **15\u201339 heavy atoms**.\n- Supported elements: `H, C, N, O, F, P, S, Cl, Br`.\n\n---\n\n## \ud83e\uddea Standardization Pipeline\n\nThe generated molecules are post-processed through the following steps:\n\n- Largest Fragment picker\n- Valence check\n- Kekulization\n- RDKit sanitization\n- Constrained Geometry optimization via **MMFF94** Molecular Dynamics\n\n---\n\n## \ud83d\udccf Evaluation Pipeline\n\nAligns and Evaluates shape similarity between generated molecules and a reference using\n**Shape Tanimoto Similarity [[3]](https://doi.org/10.1007/978-94-017-1120-3_5 )** via Gaussian Molecular Volume overlap.\n\n> Hydrogens are ignored in both reference and generated samples for this metric.\n\n---\n\n## \ud83d\udcca Performance (100 Denoising Steps)\n\n*Tested on 100,000 samples using 1,000 CCDC Virtual Screening [[4]](https://www.ccdc.cam.ac.uk/support-and-resources/downloads/) reference compounds.*\n\n### General Overview\n\n- \u23f1 **Avg time to generate 50 valid samples**: 11.46 sec (NVIDIA H100)\n- \u26a1\ufe0f **Generation speed**: 4.18 valid molecules/sec\n- \ud83d\udcbe **GPU memory (per generation thread)**: Up to 14.0 GB (`float16` 39 atoms 100 samples)\n- \ud83d\udcd0 **Avg Shape Tanimoto Similarity**: 53.32%\n- \ud83c\udfaf **Max Shape Tanimoto Similarity**: 99.69%\n- \ud83d\udd2c **Avg Chemical Tanimoto Similarity (2-hop 2048-bit Morgan Fingerprints)**: 10.87%\n- \ud83e\uddec **% Chemically novel (vs. training set)**: 99.84%\n- \u2714\ufe0f **% Valid molecules (post-standardization)**: 48%\n- \ud83d\udd01 **% Unique molecules in generated set**: 99.94%\n- \u26a1 **Average Strain (MMFF94)**: 2.36 kcal / mol\n- \ud83d\udcce **Fr\u00e9chet Fingerprint Distance (2-hop 2048-bit Morgan Fingerprints)**: \n - To ChEMBL: 4.13 \n - To PubChem: 2.64 \n - To ZINC (250k): 4.95\n\n### PoseBusters [[5]](https://doi.org/10.1039/D3SC04185A) validity check results:\n\n**Overall stats**:\n\n - PB-valid molecules: **91.33 %**\n\n**Detailed Problems**:\n\n - position: 0.01 %\n - mol_pred_loaded: 0.0 %\n - sanitization: 0.01 %\n - inchi_convertible: 0.01 %\n - all_atoms_connected: 0.0 %\n - bond_lengths: 0.24 %\n - bond_angles: 0.70 %\n - internal_steric_clash: 2.31 %\n - aromatic_ring_flatness: 3.34 %\n - non-aromatic_ring_non-flatness: 0.27 %\n\n### Synthesizability of the generated compounds\n\n#### SA Score [[6]](https://doi.org/10.1186/1758-2946-1-8)\n\n*1 (easy to make) - 10 (very difficult to make)*\n\n**Average SA Score**: **3.18**\n\n<img src=\"https://raw.githubusercontent.com/Membrizard/ml_conformer_generator/main/assets/benchmarks/sa_score_dist.png\" width=\"300\">\n\n---\n\n## Generation Examples\n\n\n\n\n\n\n---\n\n## \ud83d\udcbe Access & Licensing\n\nThe **Python package and inference code are available on GitHub** under Apache 2.0 License\n> https://github.com/Membrizard/ml_conformer_generator\n\nThe trained model **Weights** are available at\n\n> https://huggingface.co/Membrizard/ml_conformer_generator\n\nAnd are licensed under CC BY-NC-ND 4.0\n\nThe usage of the trained weights for any profit-generating activity is restricted.\n\nFor commercial licensing and inference-as-a-service, contact:\n[Denis Sapegin](https://github.com/Membrizard)\n\n---\n\n## ONNX Inference:\nFor torch Free inference an ONNX version of the model is present. \n\nWeights of the model in ONNX format are available at:\n> https://huggingface.co/Membrizard/ml_conformer_generator\n\n`egnn_chembl_15_39.onnx`\n\n`adj_mat_seer_chembl_15_39.onnx`\n\n\n```python\nfrom mlconfgen import MLConformerGeneratorONNX\nfrom rdkit import Chem\n\nmodel = MLConformerGeneratorONNX(\n egnn_onnx=\"./egnn_chembl_15_39.onnx\",\n adj_mat_seer_onnx=\"./adj_mat_seer_chembl_15_39.onnx\",\n diffusion_steps=100,\n )\n\nreference = Chem.MolFromMolFile('./assets/demo_files/yibfeu.mol')\nsamples = model.generate_conformers(reference_conformer=reference, n_samples=20, variance=2)\n\n```\nInstall ONNX GPU runtime (if needed):\n`pip install onnxruntime-gpu`\n\n---\n## Export to ONNX\nAn option to compile the model to ONNX is provided\n\nrequires `onnxscript==0.2.2`\n\n`pip install onnxscript`\n\n```python\nfrom mlconfgen import MLConformerGenerator\nfrom onnx_export import export_to_onnx\n\nmodel = MLConformerGenerator()\nexport_to_onnx(model)\n```\nThis compiles and saves the ONNX files to: `./`\n\n## Streamlit App\n\n\n\n### Running\n- Move the trained PyTorch weights into `./streamlit_app`\n\n`./streamlit_app/edm_moi_chembl_15_39.pt`\n\n`./streamlit_app/adj_mat_seer_chembl_15_39.pt`\n\n- Install the dependencies `pip install -r ./streamlit_app/requirements.txt`\n\n- Bring the app UI up:\n ```commandline\n cd ./streamlit_app\n streamlit run app.py\n ```\n\n### Streamlit App Development\n\n1. To enable development mode for the 3D viewer (`stspeck`), set `_RELEASE = False` in `./streamlit/stspeck/__init__.py`.\n\n2. Navigate to the 3D viewer frontend and start the development server:\n ```commandline\n cd ./frontend/speck/frontend\n npm run start\n ```\n \n This will launch the dev server at `http://localhost:3001`\n\n3. In a separate terminal, run the Streamlit app from the root frontend directory: \n ```commandline\n cd ./streamlit_app\n streamlit run app.py\n ```\n\n4. To build the production version of the 3D viewer, run:\n ```commandline\n cd ./streamlit_app/stspeck/frontend\n npm run build\n ```\n",
"bugtrack_url": null,
"license": null,
"summary": "Shape-constrained molecule generation via Equivariant Diffusion and GCN",
"version": "0.2.2",
"project_urls": {
"Homepage": "https://github.com/Membrizard/ml_conformer_generator",
"Issues": "https://github.com/Membrizard/ml_conformer_generator/issues"
},
"split_keywords": [
"rdkit",
" chemistry",
" diffusion",
" conformers"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "8b205dd721327c0c2b1b011f59655872e71a71bba2a62cf163cd1020e553964e",
"md5": "22edc872563dfaba3be2ca457a8a5991",
"sha256": "a4bb265816e70bd4164c023dc001c163930a56afa9771d53d8cb78906724cf72"
},
"downloads": -1,
"filename": "mlconfgen-0.2.2-py3-none-any.whl",
"has_sig": false,
"md5_digest": "22edc872563dfaba3be2ca457a8a5991",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10",
"size": 60121,
"upload_time": "2025-08-21T08:51:27",
"upload_time_iso_8601": "2025-08-21T08:51:27.222429Z",
"url": "https://files.pythonhosted.org/packages/8b/20/5dd721327c0c2b1b011f59655872e71a71bba2a62cf163cd1020e553964e/mlconfgen-0.2.2-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "39e4baa776d8ba3eef4df7f7165474493be137133fbc14ff0abf1f1e494541e9",
"md5": "359fda0d6bcc0097840b798e3263c9c8",
"sha256": "290063f06afd122b5bc07b4aaca57a76035d40aa3e7f661c0f2920f3d4d00bd6"
},
"downloads": -1,
"filename": "mlconfgen-0.2.2.tar.gz",
"has_sig": false,
"md5_digest": "359fda0d6bcc0097840b798e3263c9c8",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 50447,
"upload_time": "2025-08-21T08:51:28",
"upload_time_iso_8601": "2025-08-21T08:51:28.416737Z",
"url": "https://files.pythonhosted.org/packages/39/e4/baa776d8ba3eef4df7f7165474493be137133fbc14ff0abf1f1e494541e9/mlconfgen-0.2.2.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-08-21 08:51:28",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "Membrizard",
"github_project": "ml_conformer_generator",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "mlconfgen"
}
