<div align="center">
<img class="lightmode" style="width: 400px;" src="https://github.com/moldyn/msmhelper/blob/main/docs/logo_large_light.svg?raw=true#gh-light-mode-only" />
<p>
<a href="https://joss.theoj.org/papers/0c2a2cd2ca10b5c0bdca4d0234eb94fd">
<img src="https://joss.theoj.org/papers/0c2a2cd2ca10b5c0bdca4d0234eb94fd/status.svg" /></a>
<a href="https://github.com/wemake-services/wemake-python-styleguide" alt="wemake-python-styleguide">
<img src="https://img.shields.io/badge/style-wemake-000000.svg" /></a>
<a href="https://pypi.org/project/msmhelper" alt="PyPI">
<img src="https://img.shields.io/pypi/v/msmhelper" /></a>
<a href="https://anaconda.org/conda-forge/msmhelper" alt="conda version">
<img src="https://img.shields.io/conda/vn/conda-forge/msmhelper" /></a>
<a href="https://pepy.tech/project/msmhelper" alt="Downloads">
<img src="https://static.pepy.tech/badge/msmhelper" /></a>
<a href="https://github.com/moldyn/msmhelper/actions/workflows/pytest.yml" alt="GitHub Workflow Status">
<img src="https://img.shields.io/github/actions/workflow/status/moldyn/msmhelper/pytest.yml?branch=main"></a>
<a href="https://codecov.io/gh/moldyn/msmhelper" alt="Code coverage">
<img src="https://codecov.io/gh/moldyn/msmhelper/branch/main/graph/badge.svg?token=Ce2eW5JICI" /></a>
<a href="https://github.com/moldyn/msmhelper/actions/workflows/codeql.yml" alt="CodeQL">
<img src="https://github.com/moldyn/msmhelper/actions/workflows/codeql.yml/badge.svg?branch=main" /></a>
<a href="https://img.shields.io/pypi/pyversions/msmhelper" alt="PyPI - Python Version">
<img src="https://img.shields.io/pypi/pyversions/msmhelper" /></a>
<a href="https://moldyn.github.io/msmhelper" alt="Docs">
<img src="https://img.shields.io/badge/mkdocs-Documentation-brightgreen" /></a>
<a href="https://github.com/moldyn/msmhelper/blob/main/LICENSE" alt="License">
<img src="https://img.shields.io/github/license/moldyn/msmhelper" /></a>
</p>
<p>
<a href="https://moldyn.github.io/msmhelper">Docs</a> •
<a href="#features">Features</a> •
<a href="#installation">Installation</a> •
<a href="https://moldyn.github.io/msmhelper/faq">FAQ</a>
</p>
</div>
# msmhelper
This is a package with helper functions to work with discrete state trajectories and Markov state models. In contrast to [pyemma](https://github.com/markovmodel/PyEMMA) and [msmbuilder](https://github.com/msmbuilder/msmbuilder), it focuses on Markov state modeling based on an already existing state trajectory. Therefore, neither dimensionality reduction methods nor clustering methods are included. For a methodological overview, we recommend [Sittel and Stock](https://doi.org/10.1063/1.5049637).
This package is published in:
> **msmhelper: A Python Package for Markov State Modeling of Protein Dynamics**,
> D. Nagel, and G. Stock,
> *J. Open Source Soft.* **2023** 8 (85), 5339,
> doi: [10.21105/joss.05339](https://doi.org/10.21105/joss.05339)
We kindly ask you to cite this article in case you use this software package for published works.
## Features
- Simple usage with sleek function-based API
- High performance due to [numba](https://numba.pydata.org/)-optimized source code, checkout the [benchmark comparing to PyEMMA](https://moldyn.github.io/msmhelper/benchmark)
- [Documentation](https://moldyn.github.io/msmhelper) including tutorials
- Powerful command-line interface (CLI) to create publication-ready figures
- Supports Python 3.8-3.11
## Implemented Key Functionalities
- Hummer-Szabo projection of optimal dimensionality reduction by [Hummer and Szabo 2014](https://doi.org/10.1021/jp508375q)
- Dynamical coring by [Nagel et al. 2019](https://doi.org/10.1063/1.5081767)
- Fast extraction of pathways and MSM-based prediction of pathways based on the definition of [Nagel et al. 2020](https://pubs.acs.org/doi/10.1021/acs.jctc.0c00774)
- Fast calculation of waiting times based on both, state trajectories and MSMs
- Blazing fast [Chapman-Kolmogorov](https://www.wikiwand.com/en/Chapman%E2%80%93Kolmogorov_equation) test implementation
- Entropy-based similarity measure to compare different state discretizations, this method will be published soon in Nagel 2023
- Contact representation by [Nagel et al. 2023](https://arxiv.org/abs/2303.03814) for a compact structural representation of the states
- Command-line interface providing both, visualization and analysis methods
- Provide (non-reversible) transition matrix of all states (corresponds in pyemma to `connectivity='none', 'all'` which will (probably) [never be implemented](https://github.com/markovmodel/PyEMMA/blob/5315b8699eff2941e84577932921f694dca76f59/pyemma/msm/estimators/_msm_estimator_base.py#L110))
## Getting started
### Installation
The package is called `msmhelper` and is available via [PyPI](https://pypi.org/project/msmhelper) or [conda](https://anaconda.org/conda-forge/msmhelper). To install it, simply call:
```bash
python3 -m pip install --upgrade msmhelper
```
or
```
conda install -c conda-forge msmhelper
```
or for the latest dev version
```bash
# via ssh key
python3 -m pip install git+ssh://git@github.com/moldyn/msmhelper.git
# or via password-based login
python3 -m pip install git+https://github.com/moldyn/msmhelper.git
```
### Documentation and Tutorials
The [documentation](https://moldyn.github.io/msmhelper) serves as a comprehensive resource, offering a broad range of information such as general guidelines, API code references, and command line tool details. It also includes a Frequently Asked Questions (FAQ) section and outlines the procedures for contributing to the project.
Moreover, a suite of [tutorials](https://moldyn.github.io/msmhelper/tutorials/) is available, covering all the primary functionalities of the package. These tutorials are provided in the form of Jupyter notebooks. You can easily obtain these notebooks either directly from the [docs/tutorials](https://github.com/moldyn/msmhelper/tree/main/docs/tutorials) directory on our GitHub repository or by clicking the download buttons available on each tutorial page within the documentation.
If you prefer, you can compile the documentation on your local machine by executing the following commands:
```bash
# install all additional dependencies
python -m pip install msmhelper[docs]
# build the docs inside the site directory
python -m mkdocs build
```
### Shell Completion
Using the `bash`, `zsh` or `fish` shell click provides an easy way to
provide shell completion, checkout the
[docs](https://click.palletsprojects.com/en/8.1.x/shell-completion).
In the case of bash you need to add following line to your `~/.bashrc`
```bash
eval "$(_MSMHELPER_COMPLETE=bash_source msmhelper)"
```
In general one can call the module directly by its entry point `$ msmhelper`
or by calling the module `$ python -m msmhelper`. The latter method is
preferred to ensure using the desired python environment. For enabling
the shell completion, the entry point needs to be used.
### Usage
This package offers either a [command line interface](https://moldyn.github.io/msmhelper/reference/cli) to run standalone analysis and to create commonly-used figures, or its much more powerful [API](https://moldyn.github.io/msmhelper/tutorials/msmhelper) can be used to embedded it into an existing Python workflow. Check out the documentation for an overview over all modules and some example workflows, and for some examples see the [following section](#hummer-szabo-projection).
```python
import msmhelper as mh
# open text files
traj = mh.openmicrostates(filename, limitsfile)
# create markov state model
tmat, states = mh.estimate_markov_model(traj, lagtime=1)
...
```
### Hummer-Szabo Projection
In the following we show some sample figures produced directly with the command line tools. For more information on that, there is a [tutorial](https://moldyn.github.io/msmhelper/tutorials/hummerszabo) explaining the methods more in depth. In general we can see, that applying the HS-projection removes most projection artifacts based on coarse-graining many microstates into a few macrostates.
| Method | MSM | Hummer-Szabo MSM |
| :---: | :---: | :---: |
| Implied Timescales | [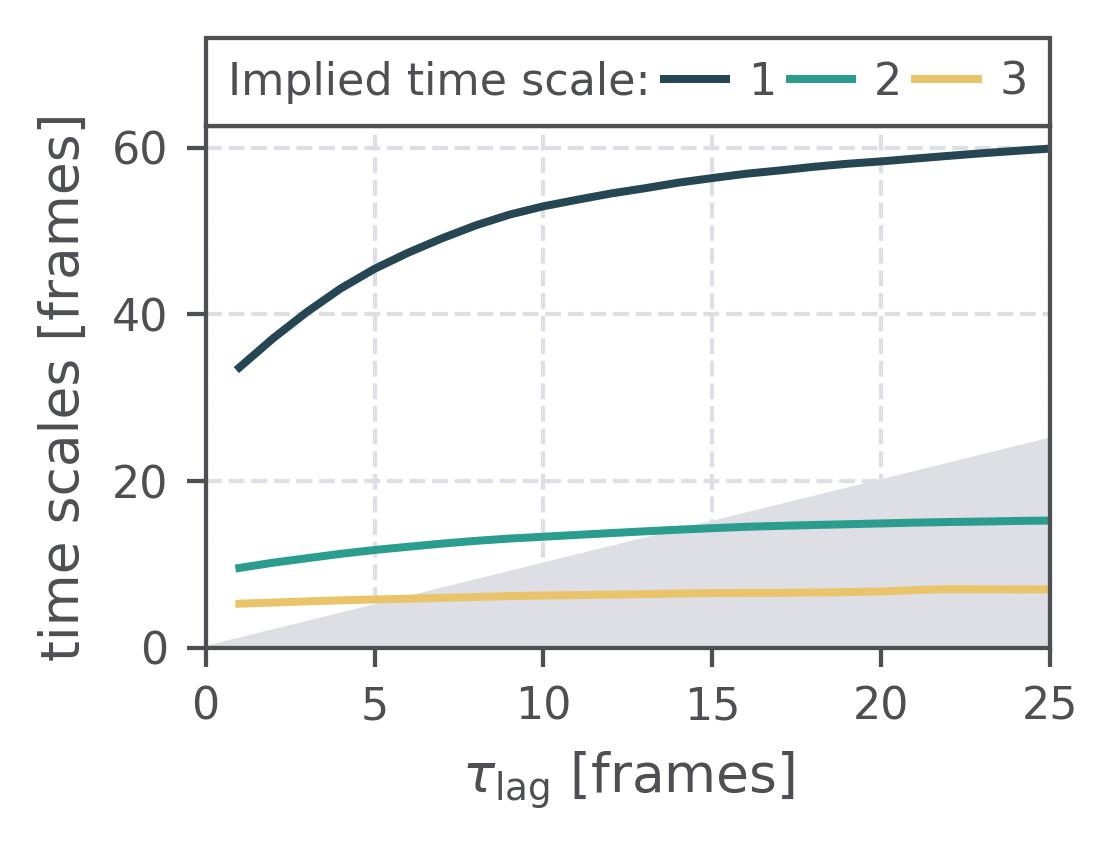](reference/cli/#msmhelper-implied-timescales) | [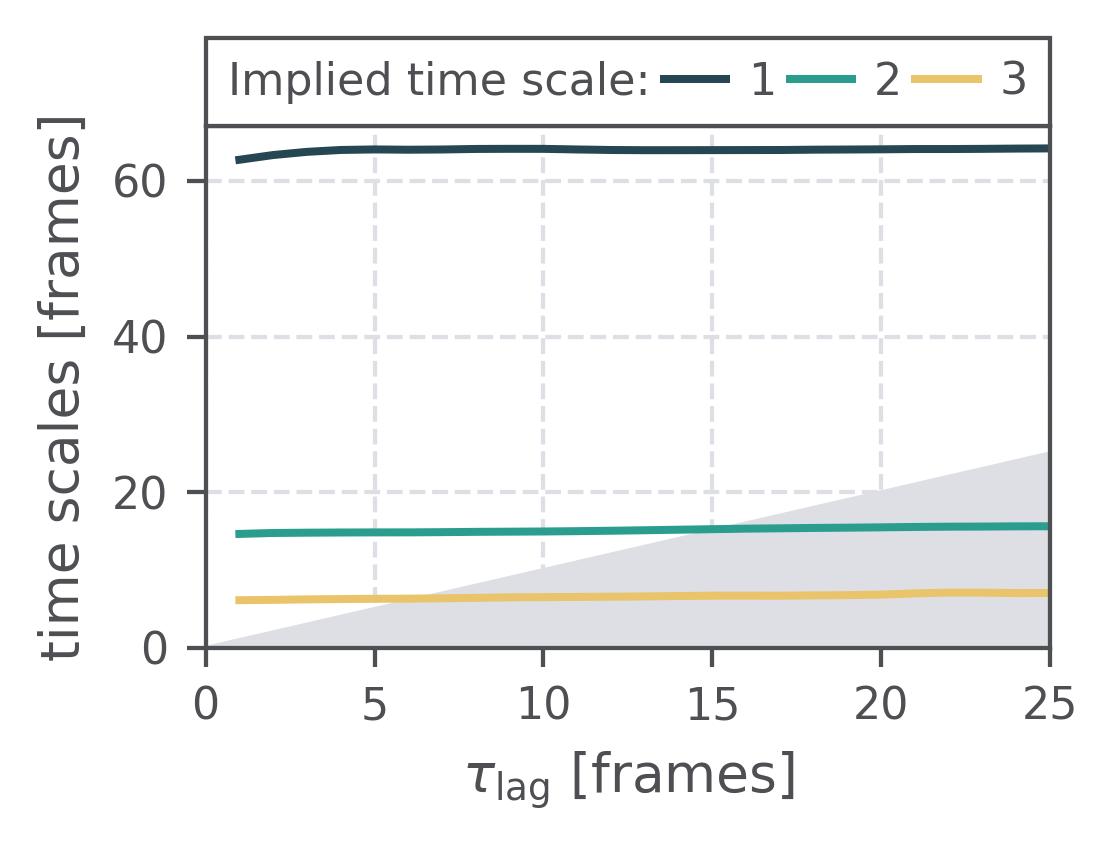](reference/cli/#msmhelper-implied-timescales) |
| Chapman-Kolmogorov test | [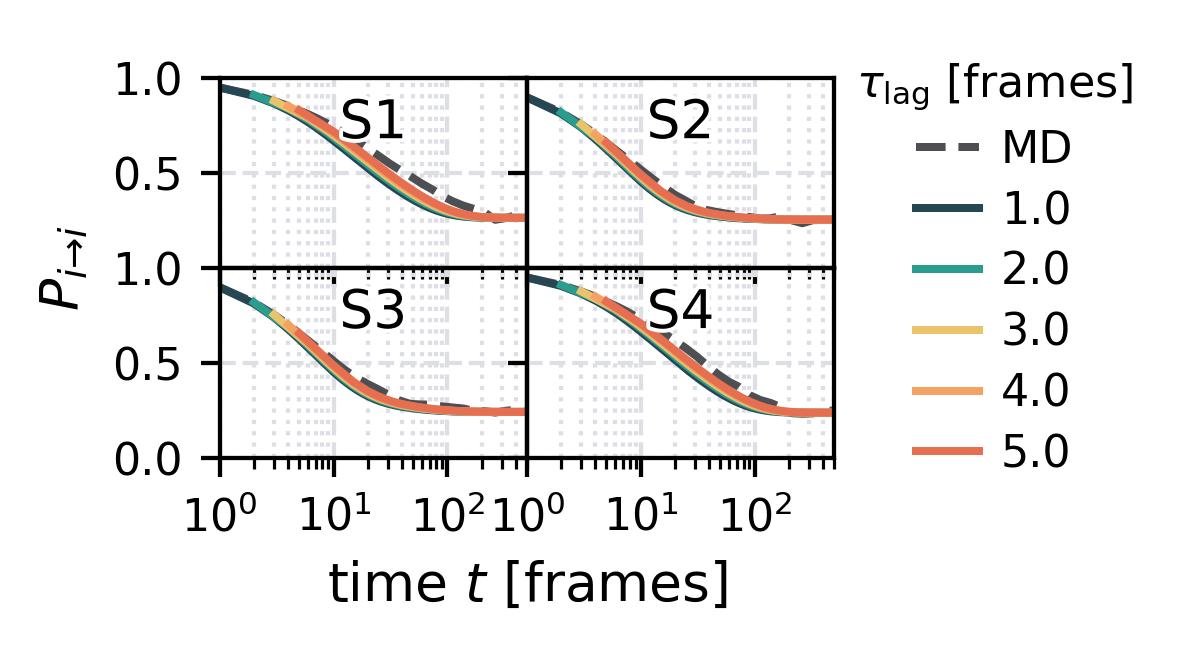](reference/cli/#msmhelper-ck-test) | [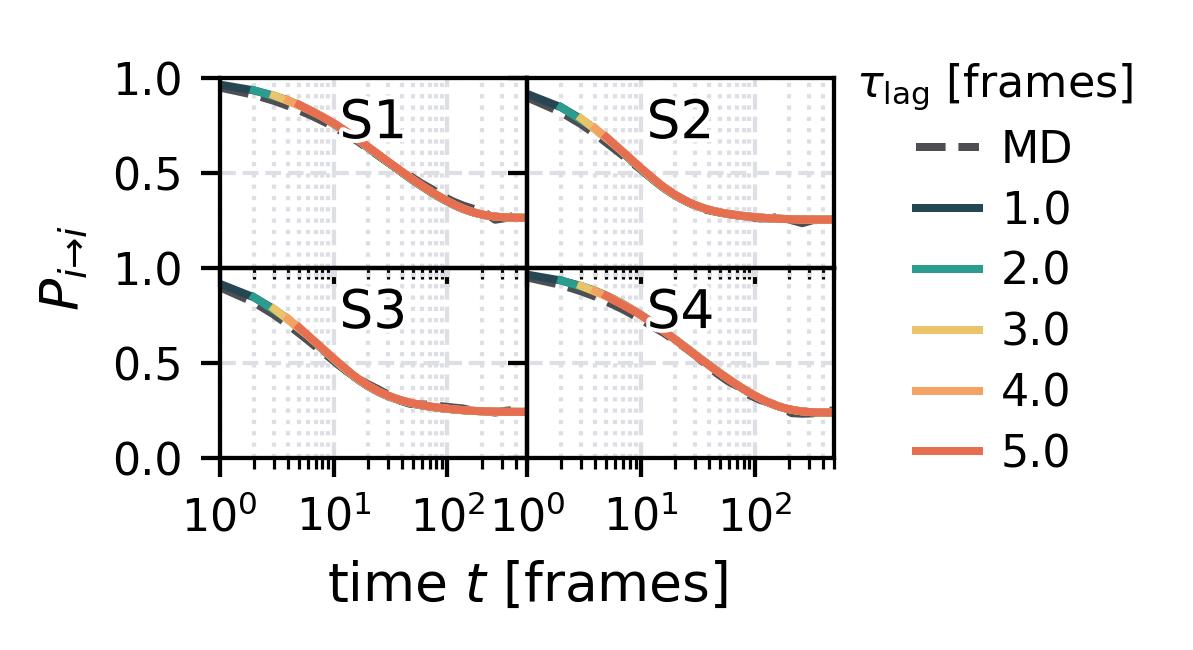](reference/cli/#msmhelper-ck-test) |
| Waiting Time Distributions | [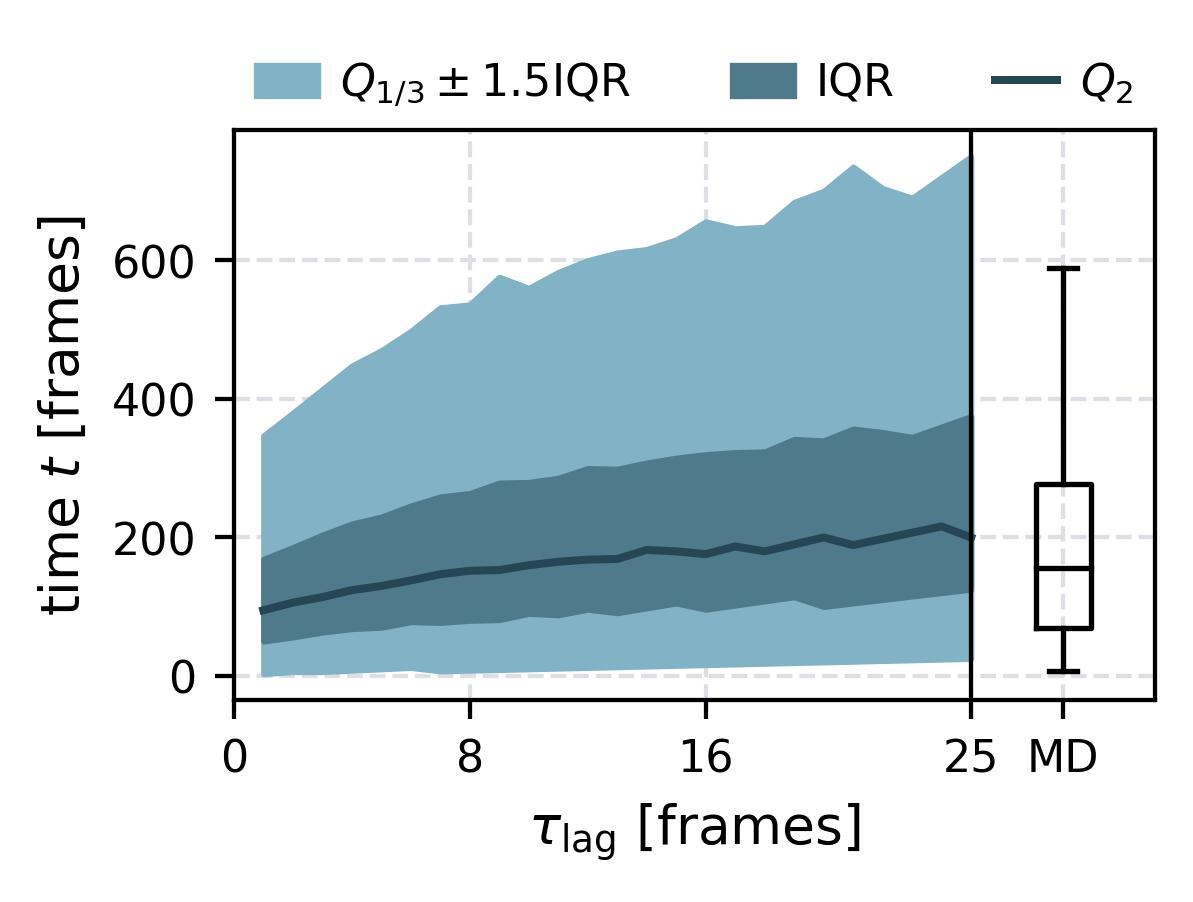](reference/cli/#msmhelper-waiting-time-dist) | [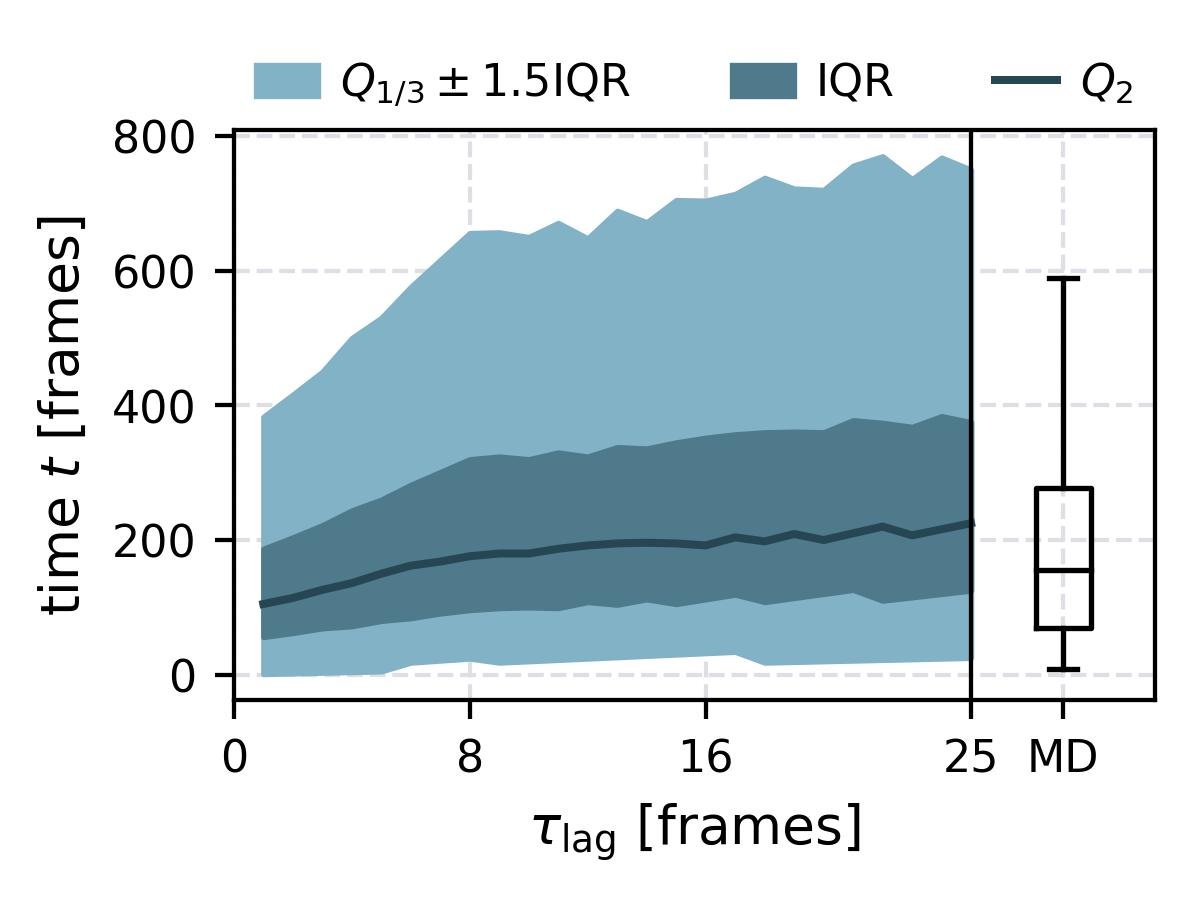](reference/cli/#msmhelper-waiting-time-dist) |
| Waiting Times | [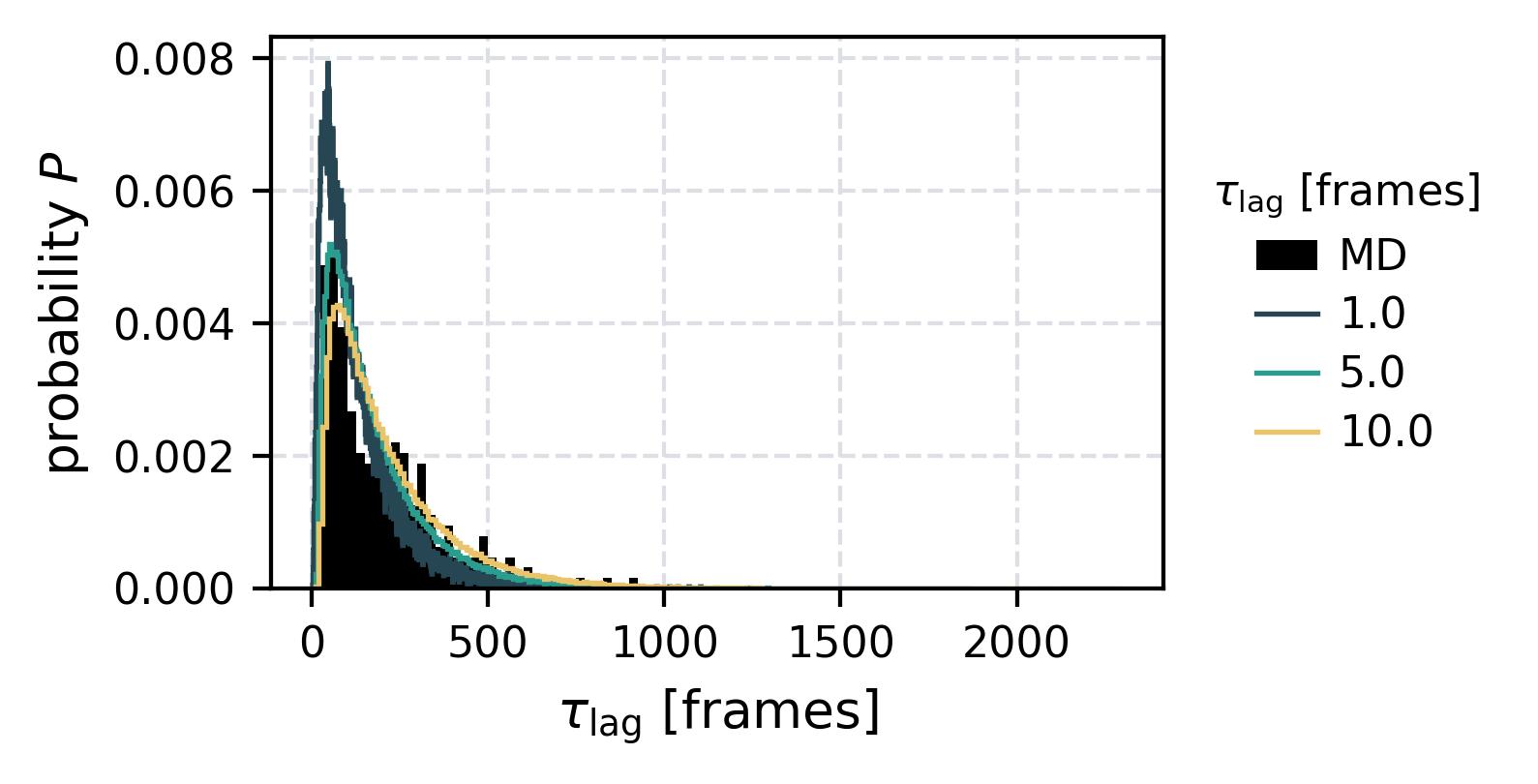](reference/cli/#msmhelper-waiting-times) | [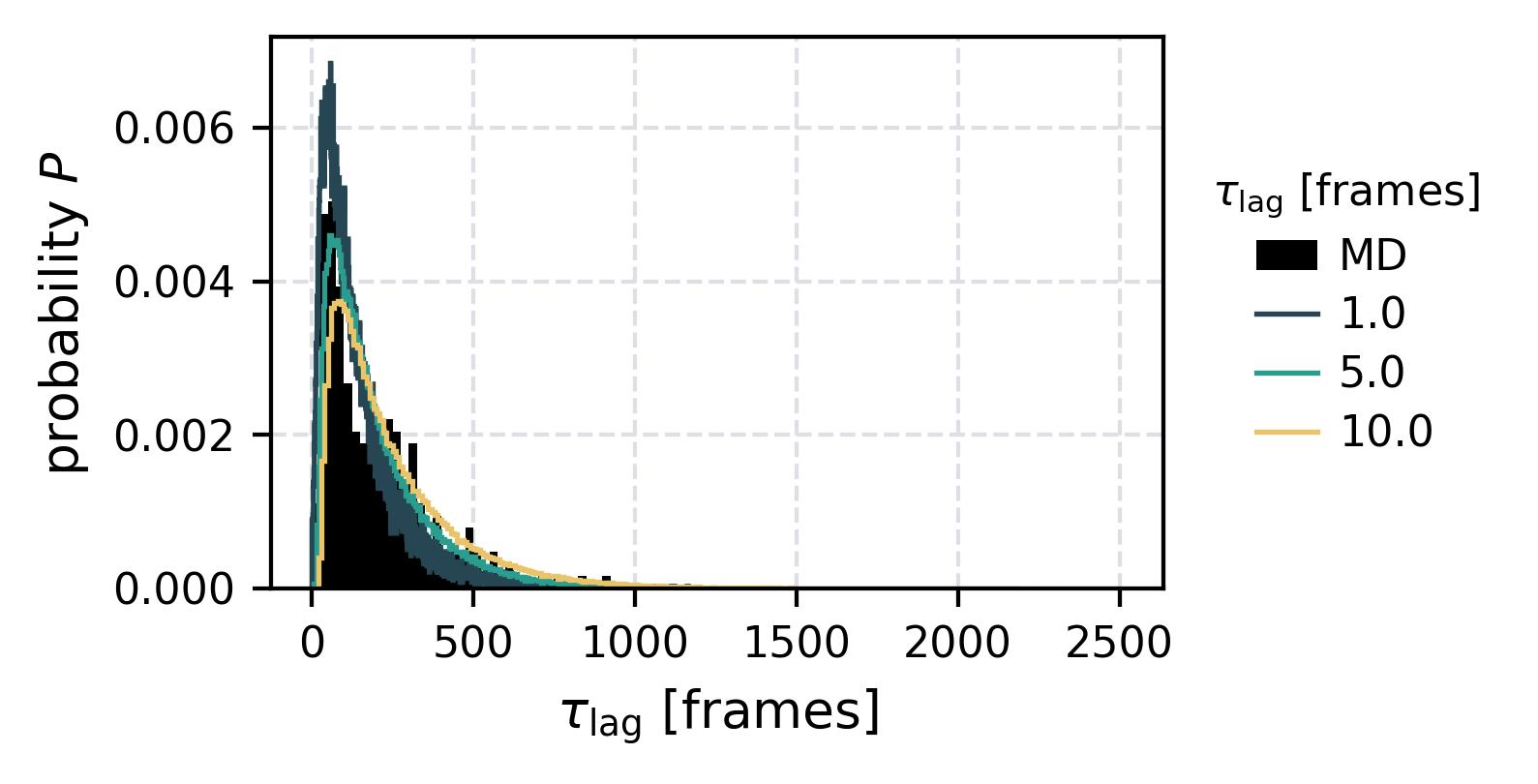](reference/cli/#msmhelper-waiting-times) |
| Contact Representation | [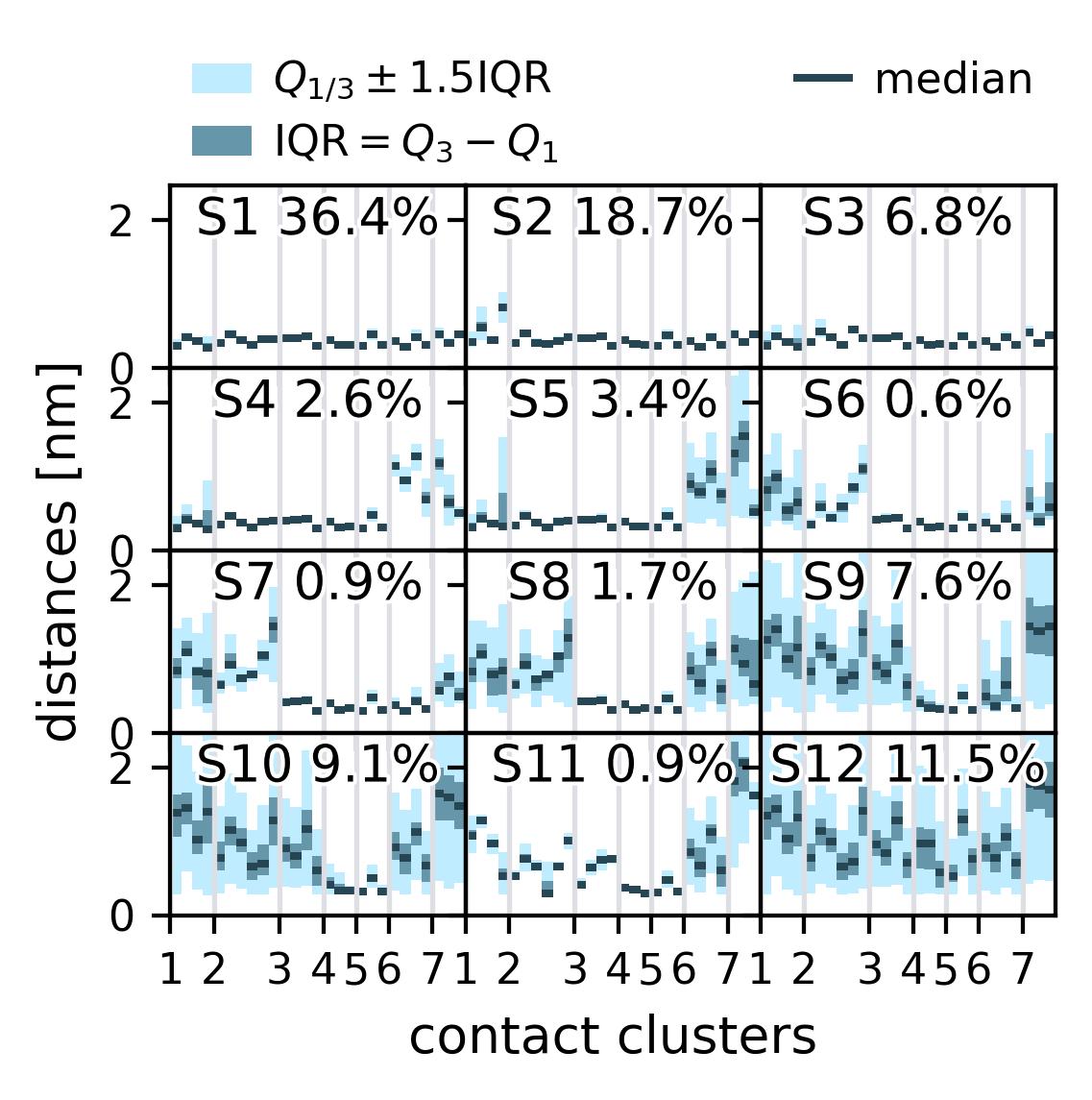](reference/cli/#msmhelper-contact-rep) | |
For more examples checkout the [tutorials](https://moldyn.github.io/msmhelper/tutorials).
## Roadmap
- Add [Buchete-Hummer test](https://doi.org/10.1021/jp0761665) as alternative for the Chapman-Kolmogorov test.
- Add a numba implementation of a parallelized autocorrelation function estimation.
- Use static type hints together with [beartype](https://github.com/beartype/beartype)
Raw data
{
"_id": null,
"home_page": "https://github.com/moldyn/msmhelper",
"name": "msmhelper",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": "",
"keywords": "MSM,Markov model,Markov state model,MD analysis",
"author": "braniii",
"author_email": "",
"download_url": "https://files.pythonhosted.org/packages/74/61/52c2695601351e2a5dd8bc82e8ec80218bd83fe84e33fc85bf2b2f1ccfa7/msmhelper-1.1.1.tar.gz",
"platform": null,
"description": "<div align=\"center\">\n \n <img class=\"lightmode\" style=\"width: 400px;\" src=\"https://github.com/moldyn/msmhelper/blob/main/docs/logo_large_light.svg?raw=true#gh-light-mode-only\" />\n\n <p>\n <a href=\"https://joss.theoj.org/papers/0c2a2cd2ca10b5c0bdca4d0234eb94fd\">\n <img src=\"https://joss.theoj.org/papers/0c2a2cd2ca10b5c0bdca4d0234eb94fd/status.svg\" /></a>\n <a href=\"https://github.com/wemake-services/wemake-python-styleguide\" alt=\"wemake-python-styleguide\">\n <img src=\"https://img.shields.io/badge/style-wemake-000000.svg\" /></a>\n <a href=\"https://pypi.org/project/msmhelper\" alt=\"PyPI\">\n <img src=\"https://img.shields.io/pypi/v/msmhelper\" /></a>\n <a href=\"https://anaconda.org/conda-forge/msmhelper\" alt=\"conda version\">\n <img src=\"https://img.shields.io/conda/vn/conda-forge/msmhelper\" /></a>\n <a href=\"https://pepy.tech/project/msmhelper\" alt=\"Downloads\">\n <img src=\"https://static.pepy.tech/badge/msmhelper\" /></a>\n <a href=\"https://github.com/moldyn/msmhelper/actions/workflows/pytest.yml\" alt=\"GitHub Workflow Status\">\n <img src=\"https://img.shields.io/github/actions/workflow/status/moldyn/msmhelper/pytest.yml?branch=main\"></a>\n <a href=\"https://codecov.io/gh/moldyn/msmhelper\" alt=\"Code coverage\">\n <img src=\"https://codecov.io/gh/moldyn/msmhelper/branch/main/graph/badge.svg?token=Ce2eW5JICI\" /></a>\n <a href=\"https://github.com/moldyn/msmhelper/actions/workflows/codeql.yml\" alt=\"CodeQL\">\n <img src=\"https://github.com/moldyn/msmhelper/actions/workflows/codeql.yml/badge.svg?branch=main\" /></a>\n <a href=\"https://img.shields.io/pypi/pyversions/msmhelper\" alt=\"PyPI - Python Version\">\n <img src=\"https://img.shields.io/pypi/pyversions/msmhelper\" /></a>\n <a href=\"https://moldyn.github.io/msmhelper\" alt=\"Docs\">\n <img src=\"https://img.shields.io/badge/mkdocs-Documentation-brightgreen\" /></a>\n <a href=\"https://github.com/moldyn/msmhelper/blob/main/LICENSE\" alt=\"License\">\n <img src=\"https://img.shields.io/github/license/moldyn/msmhelper\" /></a>\n </p>\n\n <p>\n <a href=\"https://moldyn.github.io/msmhelper\">Docs</a> \u2022\n <a href=\"#features\">Features</a> \u2022\n <a href=\"#installation\">Installation</a> \u2022\n <a href=\"https://moldyn.github.io/msmhelper/faq\">FAQ</a>\n </p>\n</div>\n\n# msmhelper\n\nThis is a package with helper functions to work with discrete state trajectories and Markov state models. In contrast to [pyemma](https://github.com/markovmodel/PyEMMA) and [msmbuilder](https://github.com/msmbuilder/msmbuilder), it focuses on Markov state modeling based on an already existing state trajectory. Therefore, neither dimensionality reduction methods nor clustering methods are included. For a methodological overview, we recommend [Sittel and Stock](https://doi.org/10.1063/1.5049637).\n\nThis package is published in:\n> **msmhelper: A Python Package for Markov State Modeling of Protein Dynamics**, \n> D. Nagel, and G. Stock, \n> *J. Open Source Soft.* **2023** 8 (85), 5339, \n> doi: [10.21105/joss.05339](https://doi.org/10.21105/joss.05339)\n\nWe kindly ask you to cite this article in case you use this software package for published works.\n\n## Features\n- Simple usage with sleek function-based API\n- High performance due to [numba](https://numba.pydata.org/)-optimized source code, checkout the [benchmark comparing to PyEMMA](https://moldyn.github.io/msmhelper/benchmark)\n- [Documentation](https://moldyn.github.io/msmhelper) including tutorials\n- Powerful command-line interface (CLI) to create publication-ready figures\n- Supports Python 3.8-3.11\n\n## Implemented Key Functionalities\n- Hummer-Szabo projection of optimal dimensionality reduction by [Hummer and Szabo 2014](https://doi.org/10.1021/jp508375q)\n- Dynamical coring by [Nagel et al. 2019](https://doi.org/10.1063/1.5081767)\n- Fast extraction of pathways and MSM-based prediction of pathways based on the definition of [Nagel et al. 2020](https://pubs.acs.org/doi/10.1021/acs.jctc.0c00774)\n- Fast calculation of waiting times based on both, state trajectories and MSMs\n- Blazing fast [Chapman-Kolmogorov](https://www.wikiwand.com/en/Chapman%E2%80%93Kolmogorov_equation) test implementation\n- Entropy-based similarity measure to compare different state discretizations, this method will be published soon in Nagel 2023\n- Contact representation by [Nagel et al. 2023](https://arxiv.org/abs/2303.03814) for a compact structural representation of the states\n- Command-line interface providing both, visualization and analysis methods\n- Provide (non-reversible) transition matrix of all states (corresponds in pyemma to `connectivity='none', 'all'` which will (probably) [never be implemented](https://github.com/markovmodel/PyEMMA/blob/5315b8699eff2941e84577932921f694dca76f59/pyemma/msm/estimators/_msm_estimator_base.py#L110))\n\n## Getting started\n### Installation\nThe package is called `msmhelper` and is available via [PyPI](https://pypi.org/project/msmhelper) or [conda](https://anaconda.org/conda-forge/msmhelper). To install it, simply call:\n```bash\npython3 -m pip install --upgrade msmhelper\n```\nor\n```\nconda install -c conda-forge msmhelper\n```\n\nor for the latest dev version\n```bash\n# via ssh key\npython3 -m pip install git+ssh://git@github.com/moldyn/msmhelper.git\n\n# or via password-based login\npython3 -m pip install git+https://github.com/moldyn/msmhelper.git\n```\n\n### Documentation and Tutorials\n\nThe [documentation](https://moldyn.github.io/msmhelper) serves as a comprehensive resource, offering a broad range of information such as general guidelines, API code references, and command line tool details. It also includes a Frequently Asked Questions (FAQ) section and outlines the procedures for contributing to the project. \nMoreover, a suite of [tutorials](https://moldyn.github.io/msmhelper/tutorials/) is available, covering all the primary functionalities of the package. These tutorials are provided in the form of Jupyter notebooks. You can easily obtain these notebooks either directly from the [docs/tutorials](https://github.com/moldyn/msmhelper/tree/main/docs/tutorials) directory on our GitHub repository or by clicking the download buttons available on each tutorial page within the documentation.\n\nIf you prefer, you can compile the documentation on your local machine by executing the following commands:\n\n```bash\n# install all additional dependencies\npython -m pip install msmhelper[docs]\n# build the docs inside the site directory\npython -m mkdocs build\n```\n\n### Shell Completion\nUsing the `bash`, `zsh` or `fish` shell click provides an easy way to\nprovide shell completion, checkout the\n[docs](https://click.palletsprojects.com/en/8.1.x/shell-completion).\nIn the case of bash you need to add following line to your `~/.bashrc`\n```bash\neval \"$(_MSMHELPER_COMPLETE=bash_source msmhelper)\"\n```\nIn general one can call the module directly by its entry point `$ msmhelper`\nor by calling the module `$ python -m msmhelper`. The latter method is\npreferred to ensure using the desired python environment. For enabling\nthe shell completion, the entry point needs to be used.\n\n### Usage\nThis package offers either a [command line interface](https://moldyn.github.io/msmhelper/reference/cli) to run standalone analysis and to create commonly-used figures, or its much more powerful [API](https://moldyn.github.io/msmhelper/tutorials/msmhelper) can be used to embedded it into an existing Python workflow. Check out the documentation for an overview over all modules and some example workflows, and for some examples see the [following section](#hummer-szabo-projection).\n```python\nimport msmhelper as mh\n\n# open text files\ntraj = mh.openmicrostates(filename, limitsfile)\n# create markov state model\ntmat, states = mh.estimate_markov_model(traj, lagtime=1)\n...\n```\n\n### Hummer-Szabo Projection\nIn the following we show some sample figures produced directly with the command line tools. For more information on that, there is a [tutorial](https://moldyn.github.io/msmhelper/tutorials/hummerszabo) explaining the methods more in depth. In general we can see, that applying the HS-projection removes most projection artifacts based on coarse-graining many microstates into a few macrostates.\n\n| Method | MSM | Hummer-Szabo MSM |\n| :---: | :---: | :---: |\n| Implied Timescales | [](reference/cli/#msmhelper-implied-timescales) | [](reference/cli/#msmhelper-implied-timescales) |\n| Chapman-Kolmogorov test | [](reference/cli/#msmhelper-ck-test) | [](reference/cli/#msmhelper-ck-test) |\n| Waiting Time Distributions | [](reference/cli/#msmhelper-waiting-time-dist) | [](reference/cli/#msmhelper-waiting-time-dist) |\n| Waiting Times | [](reference/cli/#msmhelper-waiting-times) | [](reference/cli/#msmhelper-waiting-times) |\n| Contact Representation | [](reference/cli/#msmhelper-contact-rep) | |\n\nFor more examples checkout the [tutorials](https://moldyn.github.io/msmhelper/tutorials).\n\n## Roadmap\n- Add [Buchete-Hummer test](https://doi.org/10.1021/jp0761665) as alternative for the Chapman-Kolmogorov test.\n- Add a numba implementation of a parallelized autocorrelation function estimation.\n- Use static type hints together with [beartype](https://github.com/beartype/beartype)\n",
"bugtrack_url": null,
"license": "BSD-3-Clause License",
"summary": "Helper functions for Markov State Models.",
"version": "1.1.1",
"project_urls": {
"Bug Tracker": "https://github.com/moldyn/msmhelper/issues",
"Changelog": "https://moldyn.github.io/msmhelper/changelog",
"Documentation": "https://moldyn.github.io/msmhelper",
"Homepage": "https://github.com/moldyn/msmhelper",
"Source Code": "https://github.com/moldyn/msmhelper"
},
"split_keywords": [
"msm",
"markov model",
"markov state model",
"md analysis"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "872d1375bfde7c8a49e3d28bca2f514b6724af59ccee61b20ef9c172055df0db",
"md5": "97bc67434c7b8f76ba0197210b87eb9b",
"sha256": "d43c58f4f21fb8423252322e0e82cbf00543678bb63e71945a3296cab89481dc"
},
"downloads": -1,
"filename": "msmhelper-1.1.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "97bc67434c7b8f76ba0197210b87eb9b",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 53482,
"upload_time": "2023-11-10T08:29:15",
"upload_time_iso_8601": "2023-11-10T08:29:15.466357Z",
"url": "https://files.pythonhosted.org/packages/87/2d/1375bfde7c8a49e3d28bca2f514b6724af59ccee61b20ef9c172055df0db/msmhelper-1.1.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "746152c2695601351e2a5dd8bc82e8ec80218bd83fe84e33fc85bf2b2f1ccfa7",
"md5": "09902091fceba4bb3353f4f69a499a57",
"sha256": "3c442aa49034c1d9ef6c117605c32b161192c0c679f57cdb6ca990bcae7f83a0"
},
"downloads": -1,
"filename": "msmhelper-1.1.1.tar.gz",
"has_sig": false,
"md5_digest": "09902091fceba4bb3353f4f69a499a57",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 53226,
"upload_time": "2023-11-10T08:29:17",
"upload_time_iso_8601": "2023-11-10T08:29:17.088757Z",
"url": "https://files.pythonhosted.org/packages/74/61/52c2695601351e2a5dd8bc82e8ec80218bd83fe84e33fc85bf2b2f1ccfa7/msmhelper-1.1.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-11-10 08:29:17",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "moldyn",
"github_project": "msmhelper",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [],
"lcname": "msmhelper"
}
