
Multi-Input Medical Image Machine Learning Toolkit
==================================================
The Multi-Input Medical Image Machine Learning Toolkit (MultiMedImageML) is a library of Pytorch functions that can encode multiple 3D images (designed specifically for brain images) and offer a single- or multi-label output, such as a disease detection.
Thus, with a dataset of brain images and labels, you can train a model to predict dementia or multiple sclerosis from multiple input brain images.
To install Multi Med Image ML, simply type into a standard UNIX terminal
pip install multi-med-image-ml
Overview
========
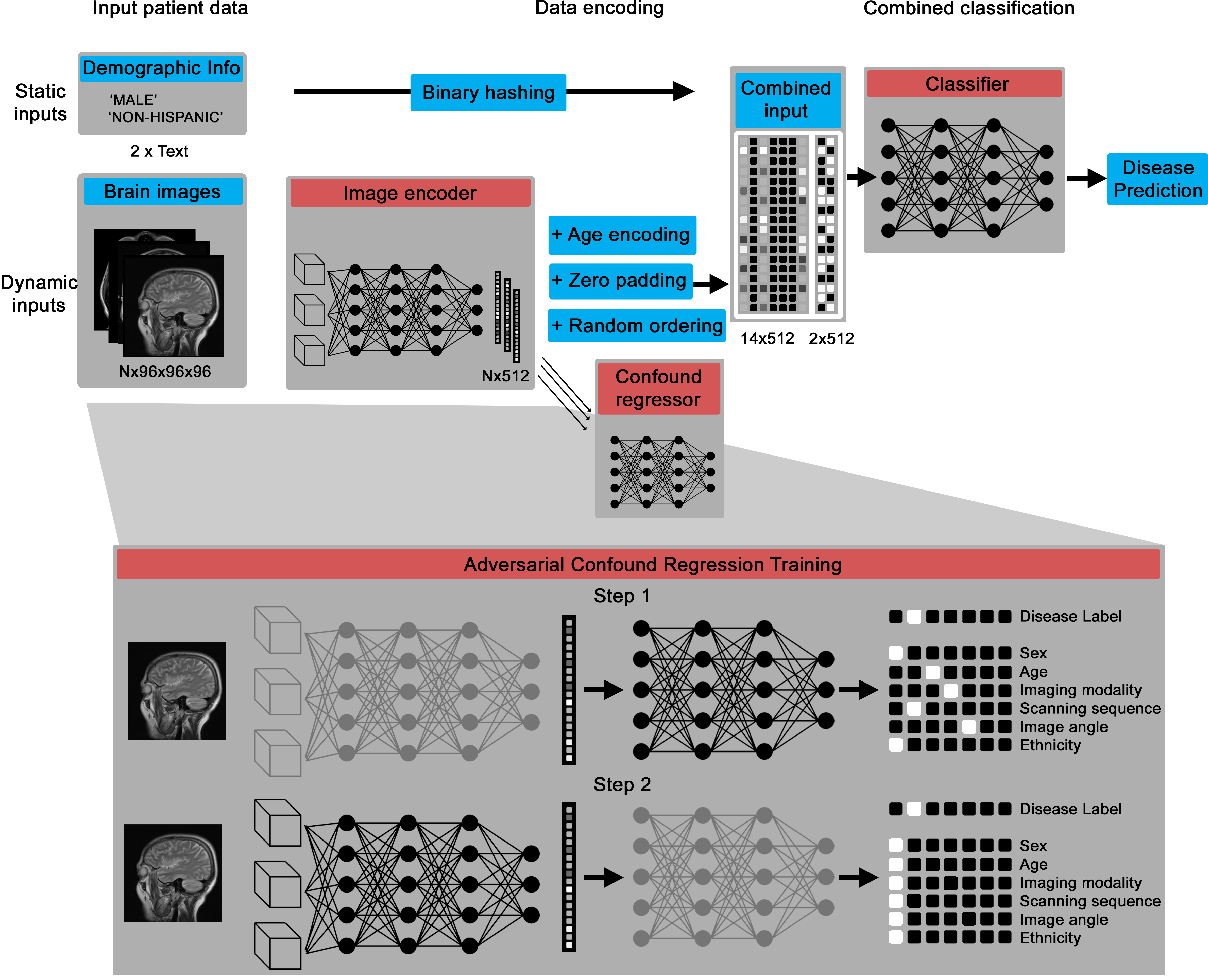
The core deep learning architecture is a Pytorch model that can take in variable numbers of 3D images (between one and 14 by default), then encodes them into a numerical vector and, through an adversarial training process, creates an intermediate representation that contains information about disease biomarkers but not confounds, like patient age and scanning site.
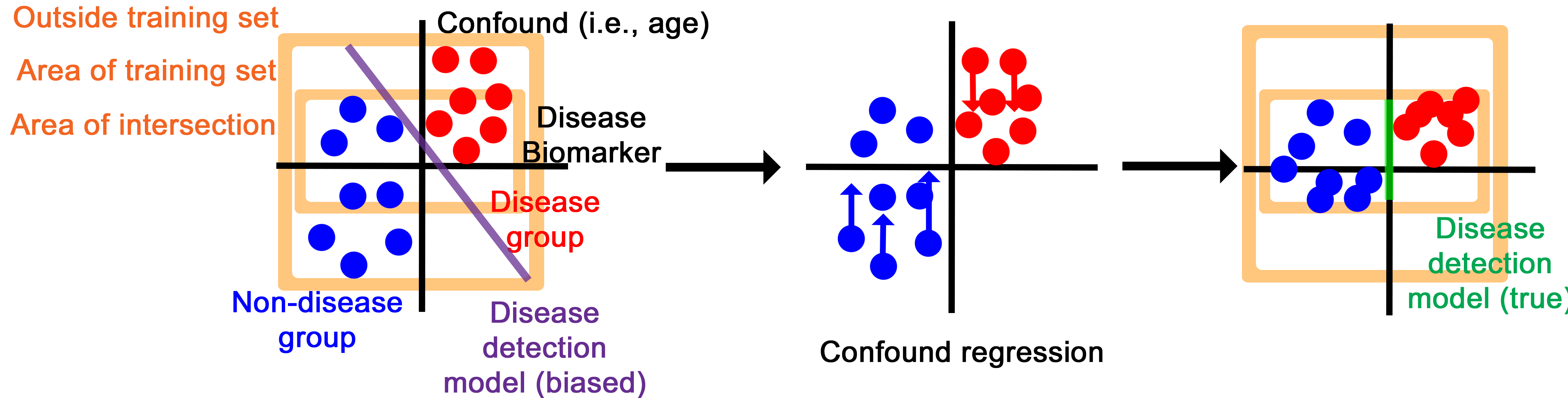
The confound regression process essentially disguises the intermediary representation to have disease biomarker features while imitating the confounding features of other groups.
Getting Started
===============
See the [Documentation](https://mleming.github.io/MultiMedImageML/build/html/).
Datasets
========
This may be used with either public benchmark datasets of brain images or internal hospital records, so long as they're represented as DICOM or NIFTI images. It was largely tested on [ADNI](https://adni.loni.usc.edu/data-samples/access-data/) and data internal to MGH. If they're represented as DICOM images, they are converted to NIFTI with metadata represented as a JSON file using [dcm2niix](https://github.com/rordenlab/dcm2niix). They may be further converted to NPY files, which are resized to a specific dimension, with the metadata represented in a pandas dataframe.
The MedImageLoader builds up this representation automatically, but it is space-intensive to do so.
Data may be represented with a folder structure.
```
.
└── control
├── 941_S_7051
│ ├── Axial_3TE_T2_STAR
│ │ └── 2022-03-07_11_03_03.0
│ │ ├── I1553008
│ │ │ ├── I1553008_Axial_3TE_T2_STAR_20220307110304_5_e3_ph.json
│ │ │ └── I1553008_Axial_3TE_T2_STAR_20220307110304_5_e3_ph.nii.gz
│ │ └── I1553014
│ │ ├── I1553014_Axial_3TE_T2_STAR_20220307110304_5_ph.json
│ │ └── I1553014_Axial_3TE_T2_STAR_20220307110304_5_ph.nii.gz
│ ├── HighResHippocampus
│ │ └── 2022-03-07_11_03_03.0
│ │ └── I1553013
│ │ ├── I1553013_HighResHippocampus_20220307110304_11.json
│ │ └── I1553013_HighResHippocampus_20220307110304_11.nii.gz
│ └── Sagittal_3D_FLAIR
│ └── 2022-03-07_11_03_03.0
│ └── I1553012
│ ├── I1553012_Sagittal_3D_FLAIR_20220307110304_3.json
│ └── I1553012_Sagittal_3D_FLAIR_20220307110304_3.nii.gz
└── 941_S_7087
├── Axial_3D_PASL__Eyes_Open_
│ └── 2022-06-15_14_38_03.0
│ └── I1591322
│ ├── I1591322_Axial_3D_PASL_(Eyes_Open)_20220615143803_6.json
│ └── I1591322_Axial_3D_PASL_(Eyes_Open)_20220615143803_6.nii.gz
└── Perfusion_Weighted
└── 2022-06-15_14_38_03.0
└── I1591323
├── I1591323_Axial_3D_PASL_(Eyes_Open)_20220615143803_7.json
└── I1591323_Axial_3D_PASL_(Eyes_Open)_20220615143803_7.nii.gz
```
In the case of the above folder structure, "/path/to/control" may simply be input into the MedImageLoader function. For multiple labels, "/path/to/test", "/path/to/test2", and so on, may also be input.
Labels and Confounds
====================
MIMIM enables for the representation of labels to classify by and confounds to regress. Confounds are represented as strings and labels can be represented as either strings or the input folder structure to MedImageLoader.
Raw data
{
"_id": null,
"home_page": null,
"name": "multi-med-image-ml",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": "Matt Leming <mleming@mgh.harvard.edu>",
"keywords": "biomedical, deep learning, ehr, machine learning, mri, pytorch",
"author": null,
"author_email": "Matt Leming <mleming@mgh.harvard.edu>",
"download_url": "https://files.pythonhosted.org/packages/cb/9f/f919cb27a106a97c4495beea5952e8cac5c154400e6483c40982d72b31fc/multi_med_image_ml-1.0.1.tar.gz",
"platform": null,
"description": "\n\nMulti-Input Medical Image Machine Learning Toolkit\n==================================================\n\nThe Multi-Input Medical Image Machine Learning Toolkit (MultiMedImageML) is a library of Pytorch functions that can encode multiple 3D images (designed specifically for brain images) and offer a single- or multi-label output, such as a disease detection.\n\nThus, with a dataset of brain images and labels, you can train a model to predict dementia or multiple sclerosis from multiple input brain images.\n\nTo install Multi Med Image ML, simply type into a standard UNIX terminal\n\n pip install multi-med-image-ml\n\n\nOverview\n========\n\n\n\nThe core deep learning architecture is a Pytorch model that can take in variable numbers of 3D images (between one and 14 by default), then encodes them into a numerical vector and, through an adversarial training process, creates an intermediate representation that contains information about disease biomarkers but not confounds, like patient age and scanning site.\n\n\n\nThe confound regression process essentially disguises the intermediary representation to have disease biomarker features while imitating the confounding features of other groups.\n\nGetting Started\n===============\n\nSee the [Documentation](https://mleming.github.io/MultiMedImageML/build/html/).\n\nDatasets\n========\n\nThis may be used with either public benchmark datasets of brain images or internal hospital records, so long as they're represented as DICOM or NIFTI images. It was largely tested on [ADNI](https://adni.loni.usc.edu/data-samples/access-data/) and data internal to MGH. If they're represented as DICOM images, they are converted to NIFTI with metadata represented as a JSON file using [dcm2niix](https://github.com/rordenlab/dcm2niix). They may be further converted to NPY files, which are resized to a specific dimension, with the metadata represented in a pandas dataframe.\n\nThe MedImageLoader builds up this representation automatically, but it is space-intensive to do so.\n\nData may be represented with a folder structure.\n\n```\n.\n\u2514\u2500\u2500 control\n \u251c\u2500\u2500 941_S_7051\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 Axial_3TE_T2_STAR\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 2022-03-07_11_03_03.0\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 I1553008\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 I1553008_Axial_3TE_T2_STAR_20220307110304_5_e3_ph.json\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1553008_Axial_3TE_T2_STAR_20220307110304_5_e3_ph.nii.gz\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1553014\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 I1553014_Axial_3TE_T2_STAR_20220307110304_5_ph.json\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1553014_Axial_3TE_T2_STAR_20220307110304_5_ph.nii.gz\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 HighResHippocampus\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 2022-03-07_11_03_03.0\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1553013\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u251c\u2500\u2500 I1553013_HighResHippocampus_20220307110304_11.json\n \u2502\u00a0\u00a0 \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1553013_HighResHippocampus_20220307110304_11.nii.gz\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 Sagittal_3D_FLAIR\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 2022-03-07_11_03_03.0\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1553012\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 I1553012_Sagittal_3D_FLAIR_20220307110304_3.json\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1553012_Sagittal_3D_FLAIR_20220307110304_3.nii.gz\n \u2514\u2500\u2500 941_S_7087\n \u251c\u2500\u2500 Axial_3D_PASL__Eyes_Open_\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 2022-06-15_14_38_03.0\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1591322\n \u2502\u00a0\u00a0 \u251c\u2500\u2500 I1591322_Axial_3D_PASL_(Eyes_Open)_20220615143803_6.json\n \u2502\u00a0\u00a0 \u2514\u2500\u2500 I1591322_Axial_3D_PASL_(Eyes_Open)_20220615143803_6.nii.gz\n \u2514\u2500\u2500 Perfusion_Weighted\n \u2514\u2500\u2500 2022-06-15_14_38_03.0\n \u2514\u2500\u2500 I1591323\n \u251c\u2500\u2500 I1591323_Axial_3D_PASL_(Eyes_Open)_20220615143803_7.json\n \u2514\u2500\u2500 I1591323_Axial_3D_PASL_(Eyes_Open)_20220615143803_7.nii.gz\n\n```\n\nIn the case of the above folder structure, \"/path/to/control\" may simply be input into the MedImageLoader function. For multiple labels, \"/path/to/test\", \"/path/to/test2\", and so on, may also be input.\n\nLabels and Confounds\n====================\n\nMIMIM enables for the representation of labels to classify by and confounds to regress. Confounds are represented as strings and labels can be represented as either strings or the input folder structure to MedImageLoader.\n",
"bugtrack_url": null,
"license": "MIT License",
"summary": "Deep learning library to encode multiple brain images and other electronic health record data in disease detection.",
"version": "1.0.1",
"project_urls": {
"Homepage": "https://github.com/mleming/MultiMedImageML"
},
"split_keywords": [
"biomedical",
" deep learning",
" ehr",
" machine learning",
" mri",
" pytorch"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "9a52711ffbea58ad83beb7c85fa4b52aea783101c8aad647f0a42297984c0b3e",
"md5": "c17e86b3278e50f61f249c1189ca5233",
"sha256": "3237ff560ef9350a06e15b3aaec1a8016e39cca79cfcbaaa6ab8955e5d608d6e"
},
"downloads": -1,
"filename": "multi_med_image_ml-1.0.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "c17e86b3278e50f61f249c1189ca5233",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 739149,
"upload_time": "2024-12-16T18:42:08",
"upload_time_iso_8601": "2024-12-16T18:42:08.482801Z",
"url": "https://files.pythonhosted.org/packages/9a/52/711ffbea58ad83beb7c85fa4b52aea783101c8aad647f0a42297984c0b3e/multi_med_image_ml-1.0.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "cb9ff919cb27a106a97c4495beea5952e8cac5c154400e6483c40982d72b31fc",
"md5": "378e05228f7b7ea13a34d5811f044fc5",
"sha256": "4cb8b32d8828c7d6f0027f9bc0009008f1ebc002bfe396146dd05e8ccc8b84b1"
},
"downloads": -1,
"filename": "multi_med_image_ml-1.0.1.tar.gz",
"has_sig": false,
"md5_digest": "378e05228f7b7ea13a34d5811f044fc5",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 4090828,
"upload_time": "2024-12-16T18:42:11",
"upload_time_iso_8601": "2024-12-16T18:42:11.679873Z",
"url": "https://files.pythonhosted.org/packages/cb/9f/f919cb27a106a97c4495beea5952e8cac5c154400e6483c40982d72b31fc/multi_med_image_ml-1.0.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-12-16 18:42:11",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "mleming",
"github_project": "MultiMedImageML",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [
{
"name": "sphinx",
"specs": []
},
{
"name": "mkdocs",
"specs": []
},
{
"name": "mkdocs-material",
"specs": []
}
],
"lcname": "multi-med-image-ml"
}
