| Name | pasmopy JSON |
| Version |
0.5.0
 JSON
JSON |
| download |
| home_page | |
| Summary | Patient-Specific Modeling in Python |
| upload_time | 2023-05-08 08:51:55 |
| maintainer | |
| docs_url | None |
| author | Hiroaki Imoto |
| requires_python | >=3.8 |
| license | Apache-2.0 |
| keywords |
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
<br>
<p align="center">
<a href="https://pasmopy.readthedocs.io/en/latest">
<img src="https://raw.githubusercontent.com/pasmopy/pasmopy/master/docs/_static/img/pasmopy-project-logo.png" width="400">
</a>
</p>
[](https://pypi.python.org/pypi/pasmopy)
[](https://github.com/pasmopy/pasmopy/actions)
[](https://pasmopy.readthedocs.io/en/latest/?badge=latest)
[](https://opensource.org/licenses/Apache-2.0)
[](https://pepy.tech/project/pasmopy)
[](https://pypi.python.org/pypi/pasmopy)
[](https://results.pre-commit.ci/latest/github/pasmopy/pasmopy/master)
[](https://github.com/psf/black)
[](https://pycqa.github.io/isort/)
[](https://doi.org/10.1016/j.isci.2022.103944)
**Pasmopy** is a scalable toolkit to identify prognostic factors for cancers based on intracellular signaling dynamics generated from personalized kinetic models. It is compatible with [biomass](https://github.com/biomass-dev/biomass) and offers the following features:
- Construction of mechanistic models from text
- Personalization of the model using transcriptome data
- Prediction of patient outcome based on _in silico_ signaling dynamics
- Sensitivity analysis for prediction of potential drug targets
## Documentation
Online documentation is available at https://pasmopy.readthedocs.io.
You can also build the documentation locally by running the following commands:
```shell
$ cd docs
$ make html
```
The site will live in `_build/html/index.html`.
## Installation
The latest stable release (and required dependencies) can be installed from [PyPI](https://pypi.python.org/pypi/pasmopy):
```
$ pip install pasmopy
```
Pasmopy requires Python 3.8+ to run.
## Example
### Building mathematical models of biochemical systems from text
This example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model with Pasmopy.
> E + S ⇄ ES → E + P
_An enzyme, E, binding to a substrate, S, to form a complex, ES, which in turn releases a product, P, regenerating the original enzyme._
```python
import os
from pasmopy import Text2Model, create_model, run_simulation
# Prepare a text file describing the biochemical reactions (e.g., `michaelis_menten.txt`)
reactions = """\
E + S ⇄ ES | kf=0.003, kr=0.001 | E=100, S=50
ES → E + P | kf=0.002
"""
observables = """
@obs Substrate: u[S]
@obs E_free: u[E]
@obs E_total: u[E] + u[ES]
@obs Product: u[P]
@obs Complex: u[ES]
"""
simulation_condition = """
@sim tspan: [0, 100]
"""
with open("michaelis_menten.txt", mode="w") as f:
f.writelines(reactions)
f.writelines(observables)
f.writelines(simulation_condition)
# Convert the text into an executable model
description = Text2Model("michaelis_menten.txt")
description.convert()
assert os.path.isdir("michaelis_menten")
# Run simulation
model = create_model("michaelis_menten")
run_simulation(model)
```
[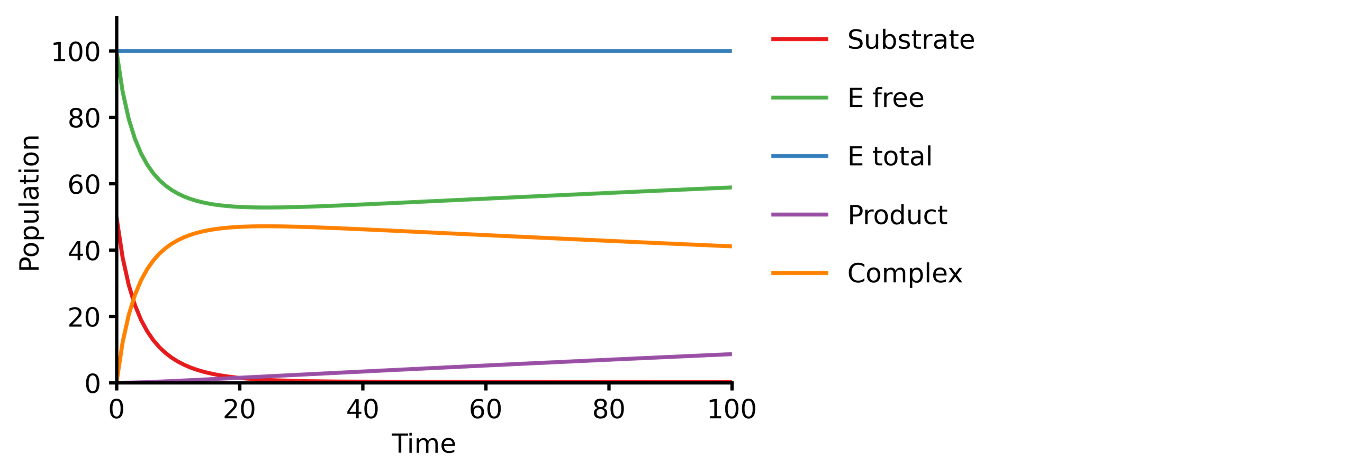](https://pasmopy.readthedocs.io/en/latest/model_development.html#michaelis-menten-enzyme-kinetics)
For more examples, please refer to the [Documentation](https://pasmopy.readthedocs.io/en/latest/).
### Personalized signaling models for cancer patient stratification
Using Pasmopy, we built a mechanistic model of ErbB receptor signaling network, trained with protein quantification data obtained from cultured cell lines, and performed _in silico_ simulation of the pathway activities on breast cancer patients using The Cancer Genome Atlas (TCGA) transcriptome datasets. The temporal activation dynamics of Akt, extracellular signal-regulated kinase (ERK), and c-Myc in each patient were able to accurately predict the difference in prognosis and sensitivity to kinase inhibitors in triple-negative breast cancer (TNBC).
For further details, please see https://pasmopy.readthedocs.io/en/latest/personalized_model.html.
## References
- Imoto, H., Yamashiro, S. & Okada, M. A text-based computational framework for patient -specific modeling for classification of cancers. _iScience_ **25**, 103944 (2022). https://doi.org/10.1016/j.isci.2022.103944
- Imoto, H., Yamashiro, S., Murakami, K. & Okada, M. Protocol for stratification of triple-negative breast cancer patients using _in silico_ signaling dynamics. _STAR Protocols_ **3**, 101619 (2022). https://doi.org/10.1016/j.xpro.2022.101619
## Author
[Hiroaki Imoto](https://github.com/himoto)
## License
[Apache License 2.0](https://github.com/pasmopy/pasmopy/blob/master/LICENSE)
Raw data
{
"_id": null,
"home_page": "",
"name": "pasmopy",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": "Hiroaki Imoto <hiroaki.imoto@ucd.ie>",
"keywords": "",
"author": "Hiroaki Imoto",
"author_email": "",
"download_url": "https://files.pythonhosted.org/packages/9a/ef/6f11af2599505d465f9b47ca0166445ca0a8f00d93563f02ab6af16e5048/pasmopy-0.5.0.tar.gz",
"platform": null,
"description": "<br>\n<p align=\"center\">\n <a href=\"https://pasmopy.readthedocs.io/en/latest\">\n <img src=\"https://raw.githubusercontent.com/pasmopy/pasmopy/master/docs/_static/img/pasmopy-project-logo.png\" width=\"400\">\n </a>\n</p>\n\n[](https://pypi.python.org/pypi/pasmopy)\n[](https://github.com/pasmopy/pasmopy/actions)\n[](https://pasmopy.readthedocs.io/en/latest/?badge=latest)\n[](https://opensource.org/licenses/Apache-2.0)\n[](https://pepy.tech/project/pasmopy)\n[](https://pypi.python.org/pypi/pasmopy)\n[](https://results.pre-commit.ci/latest/github/pasmopy/pasmopy/master)\n[](https://github.com/psf/black)\n[](https://pycqa.github.io/isort/)\n[](https://doi.org/10.1016/j.isci.2022.103944)\n\n**Pasmopy** is a scalable toolkit to identify prognostic factors for cancers based on intracellular signaling dynamics generated from personalized kinetic models. It is compatible with [biomass](https://github.com/biomass-dev/biomass) and offers the following features:\n\n- Construction of mechanistic models from text\n- Personalization of the model using transcriptome data\n- Prediction of patient outcome based on _in silico_ signaling dynamics\n- Sensitivity analysis for prediction of potential drug targets\n\n## Documentation\n\nOnline documentation is available at https://pasmopy.readthedocs.io.\n\nYou can also build the documentation locally by running the following commands:\n\n```shell\n$ cd docs\n$ make html\n```\n\nThe site will live in `_build/html/index.html`.\n\n## Installation\n\nThe latest stable release (and required dependencies) can be installed from [PyPI](https://pypi.python.org/pypi/pasmopy):\n\n```\n$ pip install pasmopy\n```\n\nPasmopy requires Python 3.8+ to run.\n\n## Example\n\n### Building mathematical models of biochemical systems from text\n\nThis example shows you how to build a simple Michaelis-Menten two-step enzyme catalysis model with Pasmopy.\n\n> E + S \u21c4 ES \u2192 E + P\n\n_An enzyme, E, binding to a substrate, S, to form a complex, ES, which in turn releases a product, P, regenerating the original enzyme._\n\n```python\nimport os\nfrom pasmopy import Text2Model, create_model, run_simulation\n\n# Prepare a text file describing the biochemical reactions (e.g., `michaelis_menten.txt`)\nreactions = \"\"\"\\\nE + S \u21c4 ES | kf=0.003, kr=0.001 | E=100, S=50\nES \u2192 E + P | kf=0.002\n\"\"\"\n\nobservables = \"\"\"\n@obs Substrate: u[S]\n@obs E_free: u[E]\n@obs E_total: u[E] + u[ES]\n@obs Product: u[P]\n@obs Complex: u[ES]\n\"\"\"\n\nsimulation_condition = \"\"\"\n@sim tspan: [0, 100]\n\"\"\"\n\nwith open(\"michaelis_menten.txt\", mode=\"w\") as f:\n f.writelines(reactions)\n f.writelines(observables)\n f.writelines(simulation_condition)\n\n# Convert the text into an executable model\ndescription = Text2Model(\"michaelis_menten.txt\")\ndescription.convert()\nassert os.path.isdir(\"michaelis_menten\")\n\n# Run simulation\nmodel = create_model(\"michaelis_menten\")\nrun_simulation(model)\n```\n\n[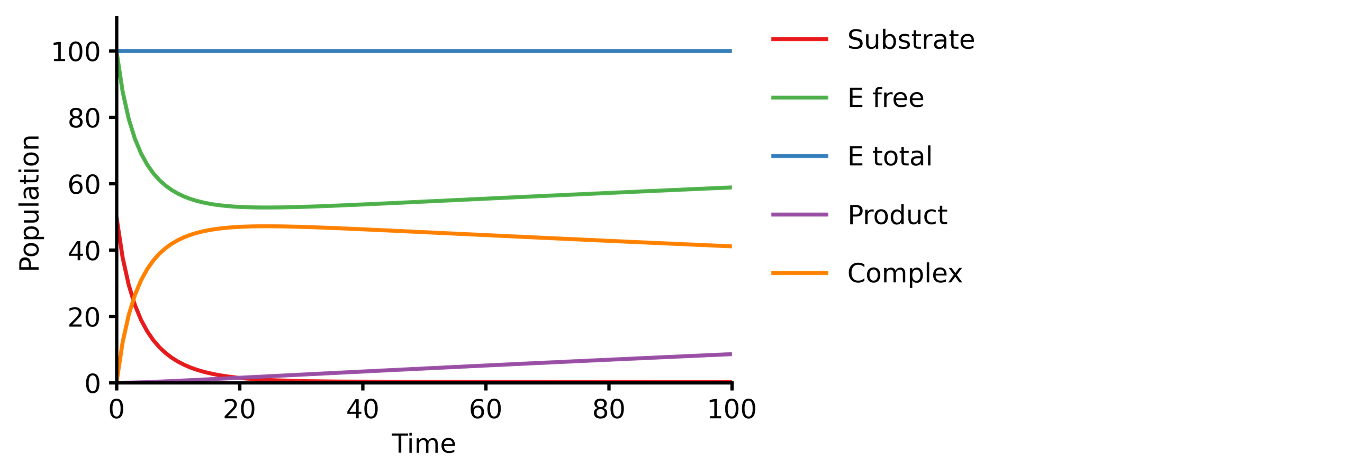](https://pasmopy.readthedocs.io/en/latest/model_development.html#michaelis-menten-enzyme-kinetics)\n\nFor more examples, please refer to the [Documentation](https://pasmopy.readthedocs.io/en/latest/).\n\n### Personalized signaling models for cancer patient stratification\n\nUsing Pasmopy, we built a mechanistic model of ErbB receptor signaling network, trained with protein quantification data obtained from cultured cell lines, and performed _in silico_ simulation of the pathway activities on breast cancer patients using The Cancer Genome Atlas (TCGA) transcriptome datasets. The temporal activation dynamics of Akt, extracellular signal-regulated kinase (ERK), and c-Myc in each patient were able to accurately predict the difference in prognosis and sensitivity to kinase inhibitors in triple-negative breast cancer (TNBC).\n\nFor further details, please see https://pasmopy.readthedocs.io/en/latest/personalized_model.html.\n\n## References\n\n- Imoto, H., Yamashiro, S. & Okada, M. A text-based computational framework for patient -specific modeling for classification of cancers. _iScience_ **25**, 103944 (2022). https://doi.org/10.1016/j.isci.2022.103944\n\n- Imoto, H., Yamashiro, S., Murakami, K. & Okada, M. Protocol for stratification of triple-negative breast cancer patients using _in silico_ signaling dynamics. _STAR Protocols_ **3**, 101619 (2022). https://doi.org/10.1016/j.xpro.2022.101619\n\n## Author\n\n[Hiroaki Imoto](https://github.com/himoto)\n\n## License\n\n[Apache License 2.0](https://github.com/pasmopy/pasmopy/blob/master/LICENSE)\n",
"bugtrack_url": null,
"license": "Apache-2.0",
"summary": "Patient-Specific Modeling in Python",
"version": "0.5.0",
"project_urls": {
"bugtracker": "https://github.com/pasmopy/pasmopy/issues",
"documentation": "https://pasmopy.readthedocs.io/en/latest/",
"repository": "https://github.com/pasmopy/pasmopy"
},
"split_keywords": [],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "19a90d5a584980bbad35f9ba556a44a45c1dd87a468f40c88fd9016254f5c9b5",
"md5": "83cda67713ce8b7d0a9d1a76d8e3d56d",
"sha256": "57a4a4b38af13a1cb1700ab293140344746cc609a40008fa6a6874f85628b42d"
},
"downloads": -1,
"filename": "pasmopy-0.5.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "83cda67713ce8b7d0a9d1a76d8e3d56d",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 19385,
"upload_time": "2023-05-08T08:51:53",
"upload_time_iso_8601": "2023-05-08T08:51:53.916127Z",
"url": "https://files.pythonhosted.org/packages/19/a9/0d5a584980bbad35f9ba556a44a45c1dd87a468f40c88fd9016254f5c9b5/pasmopy-0.5.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "9aef6f11af2599505d465f9b47ca0166445ca0a8f00d93563f02ab6af16e5048",
"md5": "578ff3b378a723dc89f6a6c0842365fc",
"sha256": "a9f8cf8bd03fa2f995e6232d347b3105e8ea6466c86d89f8e54db8edd5c0df18"
},
"downloads": -1,
"filename": "pasmopy-0.5.0.tar.gz",
"has_sig": false,
"md5_digest": "578ff3b378a723dc89f6a6c0842365fc",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 22461,
"upload_time": "2023-05-08T08:51:55",
"upload_time_iso_8601": "2023-05-08T08:51:55.729484Z",
"url": "https://files.pythonhosted.org/packages/9a/ef/6f11af2599505d465f9b47ca0166445ca0a8f00d93563f02ab6af16e5048/pasmopy-0.5.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-05-08 08:51:55",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "pasmopy",
"github_project": "pasmopy",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "pasmopy"
}
