# Pyggseqlogo
Python version of [ggseqlogo](https://github.com/omarwagih/ggseqlogo). Based on [plotnine](https://github.com/has2k1/plotnine/) (Python version of ggplot2). A derivative of [plotnineSeqSuite](https://github.com/caotianze/plotnineseqsuite/).
## Cite this work
Cao, T., Li, Q., Huang, Y. et al. plotnineSeqSuite: a Python package for visualizing sequence data using ggplot2 style. BMC Genomics 24, 585 (2023). https://doi.org/10.1186/s12864-023-09677-8
## Installation
`pip install pyggseqlogo`
## Getting Started
```Python
from plotnine import ggplot
from plotnineseqsuite import geom_logo
from plotnineseqsuite.data import seqs_dna
from pyggseqlogo import ggseqlogo, theme_logo
```
The code is based on plotnineSeqSuite.
```Python
ggplot() + geom_logo(data=seqs_dna['MA0001.1']) + theme_logo()
```
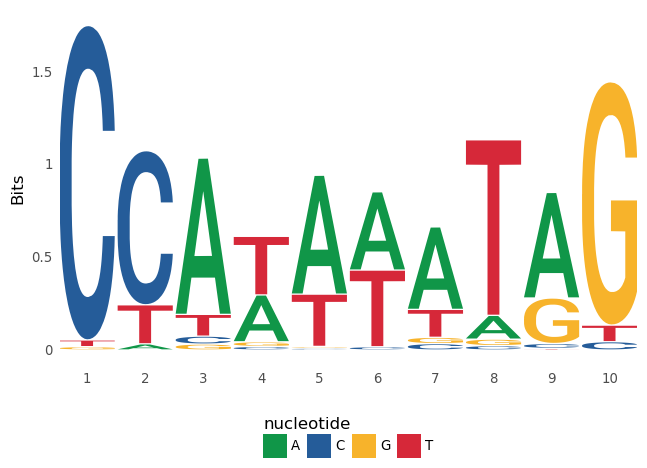
Pyggseqlogo provides a wrapper function for the above code.
```Python
ggseqlogo(seqs_dna['MA0001.1'])
```
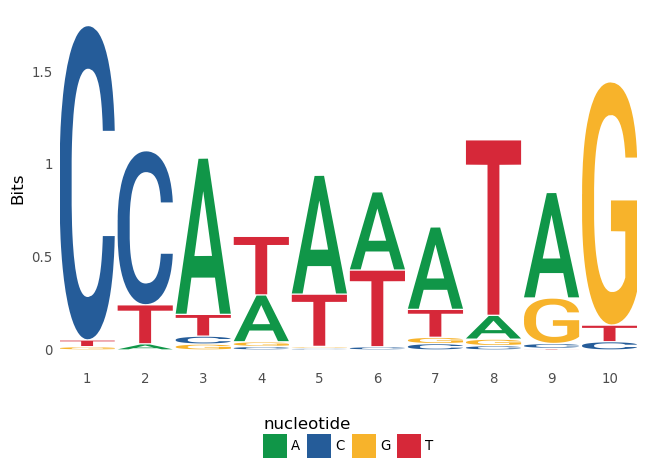
facet_wrap
```Python
ggseqlogo(seqs_dna, ncol=4)
```
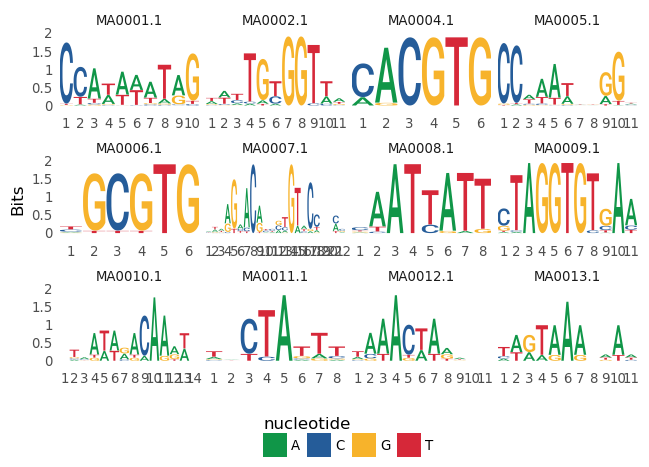
## Where to go next
If you want to draw more complex sequence logos or other sequence-related diagrams, we recommend visiting the [plotnineSeqSuite homepage](https://github.com/caotianze/plotnineseqsuite/) for details.
Raw data
{
"_id": null,
"home_page": "https://github.com/caotianze/pyggseqlogo",
"name": "pyggseqlogo",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.11",
"maintainer_email": null,
"keywords": "ggplot2, plotnine, ggseqlogo, Sequence logo",
"author": "Cao Tianze",
"author_email": "hnrcao@qq.com",
"download_url": "https://files.pythonhosted.org/packages/e5/ef/05dcbef4c5c6d32678131884324272f864a88fd6bea5bd3ea3611d56d68e/pyggseqlogo-1.1.2.tar.gz",
"platform": null,
"description": "# Pyggseqlogo\r\nPython version of [ggseqlogo](https://github.com/omarwagih/ggseqlogo). Based on [plotnine](https://github.com/has2k1/plotnine/) (Python version of ggplot2). A derivative of [plotnineSeqSuite](https://github.com/caotianze/plotnineseqsuite/).\r\n## Cite this work\r\nCao, T., Li, Q., Huang, Y. et al. plotnineSeqSuite: a Python package for visualizing sequence data using ggplot2 style. BMC Genomics 24, 585 (2023). https://doi.org/10.1186/s12864-023-09677-8\r\n## Installation\r\n`pip install pyggseqlogo`\r\n## Getting Started\r\n```Python\r\nfrom plotnine import ggplot\r\nfrom plotnineseqsuite import geom_logo\r\nfrom plotnineseqsuite.data import seqs_dna\r\nfrom pyggseqlogo import ggseqlogo, theme_logo\r\n```\r\nThe code is based on plotnineSeqSuite.\r\n```Python\r\nggplot() + geom_logo(data=seqs_dna['MA0001.1']) + theme_logo()\r\n```\r\n \r\nPyggseqlogo provides a wrapper function for the above code.\r\n```Python\r\nggseqlogo(seqs_dna['MA0001.1'])\r\n```\r\n \r\nfacet_wrap\r\n```Python\r\nggseqlogo(seqs_dna, ncol=4)\r\n```\r\n\r\n## Where to go next\r\nIf you want to draw more complex sequence logos or other sequence-related diagrams, we recommend visiting the [plotnineSeqSuite homepage](https://github.com/caotianze/plotnineseqsuite/) for details.\r\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Python version of ggseqlogo. Based on plotnine (Python version of ggplot2). A derivative of plotnineSeqSuite.",
"version": "1.1.2",
"project_urls": {
"Bug Tracker": "https://github.com/caotianze/pyggseqlogo/issues",
"Homepage": "https://github.com/caotianze/pyggseqlogo"
},
"split_keywords": [
"ggplot2",
" plotnine",
" ggseqlogo",
" sequence logo"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "e5ef05dcbef4c5c6d32678131884324272f864a88fd6bea5bd3ea3611d56d68e",
"md5": "6e11f67175d182c40c303e77004ac734",
"sha256": "eb475ab8911d4c574b94c8919f768c05164e7206d00e5a7213aca99f65f50e3e"
},
"downloads": -1,
"filename": "pyggseqlogo-1.1.2.tar.gz",
"has_sig": false,
"md5_digest": "6e11f67175d182c40c303e77004ac734",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.11",
"size": 3516,
"upload_time": "2024-12-13T05:59:10",
"upload_time_iso_8601": "2024-12-13T05:59:10.186965Z",
"url": "https://files.pythonhosted.org/packages/e5/ef/05dcbef4c5c6d32678131884324272f864a88fd6bea5bd3ea3611d56d68e/pyggseqlogo-1.1.2.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-12-13 05:59:10",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "caotianze",
"github_project": "pyggseqlogo",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "pyggseqlogo"
}
