[](https://pypi.python.org/pypi/pygmmis/)
[](https://github.com/pmelchior/pygmmis/blob/master/LICENSE.md)
[](https://doi.org/10.1016/j.ascom.2018.09.013)
[](http://arxiv.org/abs/1611.05806)
# pyGMMis
Need a simple and powerful Gaussian-mixture code in pure python? It can be as easy as this:
```python
import pygmmis
gmm = pygmmis.GMM(K=K, D=D) # K components, D dimensions
logL, U = pygmmis.fit(gmm, data) # logL = log-likelihood, U = association of data to components
```
However, **pyGMMis** has a few extra tricks up its sleeve.
* It can account for independent multivariate normal measurement errors for each of the observed samples, and then recovers an estimate of the error-free distribution. This technique is known as "Extreme Deconvolution" by Bovy, Hogg & Roweis (2011).
* It works with missing data (features) by setting the respective elements of the covariance matrix to a vary large value, thus effectively setting the weights of the missing feature to 0.
* It can deal with gaps (aka "truncated data") and variable sample completeness as long as
* you know the incompleteness over the entire feature space,
* and the incompleteness does not depend on the sample density (missing at random).
* It can incorporate a "background" distribution (implemented is a uniform one) and separate signal from background, with the former being fit by the GMM.
* It keeps track of which components need to be evaluated in which regions of the feature space, thereby substantially increasing the performance for fragmented data.
If you want more context and details on those capabilities, have a look at this [blog post](http://pmelchior.net/blog/gaussian-mixture-models-for-astronomy.html).
Under the hood, **pyGMMis** uses the Expectation-Maximization procedure. When dealing with sample incompleteness it generates its best guess of the unobserved samples on the fly given the current model fit to the observed samples.
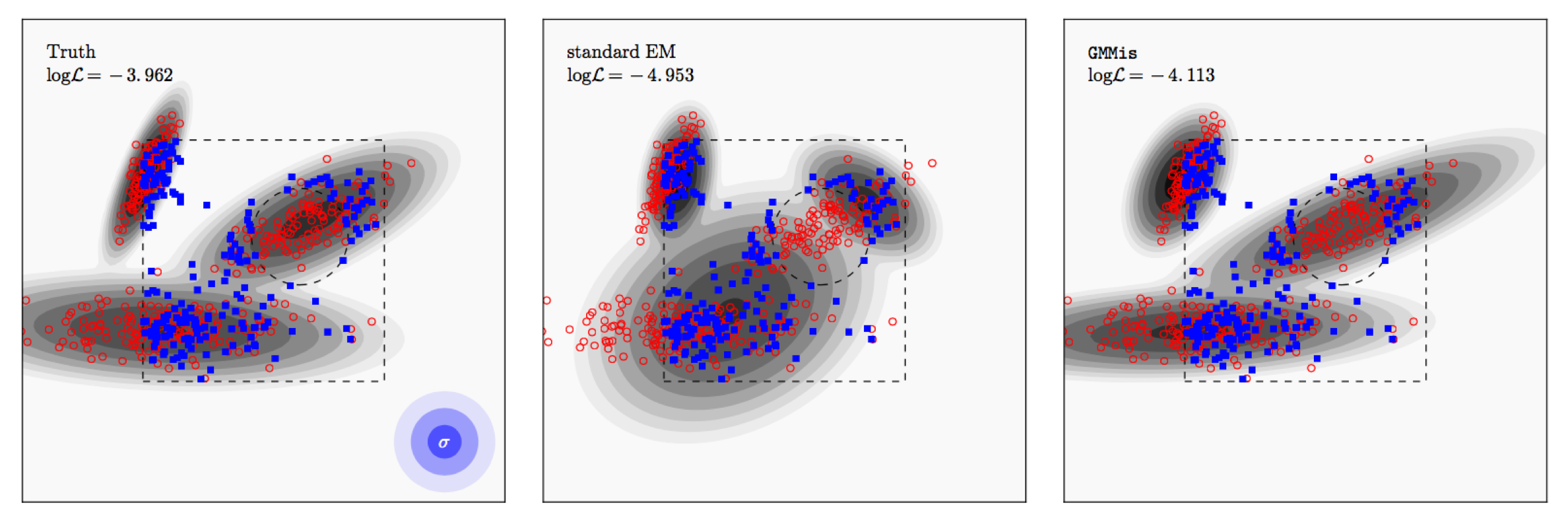
In the example above, the true distribution is shown as contours in the left panel. We then draw 400 samples from it (red), add Gaussian noise to them (1,2,3 sigma contours shown in blue), and select only samples within the box but outside of the circle (blue).
The code is written in pure python (developed and tested in 2.7), parallelized with `multiprocessing`, and is capable of performing density estimation with millions of samples and thousands of model components on machines with sufficient memory.
More details are in the paper listed in the file `CITATION.cff`.
## Installation and Prerequisites
You can either clone the repo and install by `python setup.py install` or get the latest release with
```
pip install pygmmis
```
Dependencies:
* numpy
* scipy
* multiprocessing
* parmap
## How to run the code
1. Create a GMM object with the desired component number K and data dimensionality D:
```gmm = pygmmis.GMM(K=K, D=D) ```
3. Define a callback for the completeness function. When called with with `data` with shape `(N,D)` and returns the probability of each sample getting observed. Two simple examples:
```python
def cutAtSix(coords):
"""Selects all samples whose first coordinate is < 6"""
return (coords[:,0] < 6)
def selSlope(coords, rng=np.random):
"""Selects probabilistically according to first coordinate x:
Omega = 1 for x < 0
= 1-x for x = 0 .. 1
= 0 for x > 1
"""
return np.max(0, np.min(1, 1 - coords[:,0]))
```
4. If the samples are noisy (i.e. they have positional uncertainties), you need to provide the covariance matrix of each data sample, or one for all in case of i.i.d. noise.
4. If the samples are noisy *and* there completeness function isn't constant, you need to provide a callback function that returns an estimate of the covariance at arbitrary locations:
```python
# example 1: simply using the same covariance for all samples
dispersion = 1
default_covar = np.eye(D) * dispersion**2
covar_cb = lambda coords: default_covar
# example: use the covariance of the nearest neighbor.
def covar_tree_cb(coords, tree, covar):
"""Return the covariance of the nearest neighbor of coords in data."""
dist, ind = tree.query(coords, k=1)
return covar[ind.flatten()]
from sklearn.neighbors import KDTree
tree = KDTree(data, leaf_size=100)
from functools import partial
covar_cb = partial(covar_tree_cb, tree=tree, covar=covar)
```
5. If there is a uniform background signal, you need to define it. Because a uniform distribution is normalizable only if its support is finite, you need to decide on the footprint over which the background model is present, e.g.:
```python
footprint = data.min(axis=0), data.max(axis=0)
amp = 0.3
bg = pygmmis.Background(footprint, amp=amp)
# fine tuning, if desired
bg.amp_min = 0.1
bg.amp_max = 0.5
bg.adjust_amp = False # freezes bg.amp at current value
```
6. Select an initialization method. This tells the GMM what initial parameters is should assume. The options are `'minmax','random','kmeans','none'`. See the respective functions for details:
* `pygmmis.initFromDataMinMax()`
* `pygmmis.initFromDataAtRandom()`
* `pygmmis.initFromKMeans()`
For difficult situations, or if you are not happy with the convergence, you may want to experiment with your own initialization. All you have to do is set `gmm.amp`, `gmm.mean`, and `gmm.covar` to desired values and use `init_method='none'`.
7. Decide to freeze out any components. This makes sense if you *know* some of the parameters of the components. You can freeze amplitude, mean, or covariance of any component by listing them in a dictionary, e.g:
```python
frozen={"amp": [1,2], "mean": [], "covar": [1]}
```
This freezes the amplitudes of component 1 and 2 (NOTE: Counting starts at 0), and the covariance of 1.
8. Run the fitter:
```python
w = 0.1 # minimum covariance regularization, same units as data
cutoff = 5 # segment the data set into neighborhood within 5 sigma around components
tol = 1e-3 # tolerance on logL to terminate EM
# define RNG for deterministic behavior
from numpy.random import RandomState
seed = 42
rng = RandomState(seed)
# run EM
logL, U = pygmmis.fit(gmm, data, init_method='random',\
sel_callback=cb, covar_callback=covar_cb, w=w, cutoff=cutoff,\
background=bg, tol=tol, frozen=frozen, rng=rng)
```
This runs the EM procedure until tolerance is reached and returns the final mean log-likelihood of all samples, and the neighborhood of each component (indices of data samples that are within cutoff of a GMM component).
9. Evaluate the model:
```python
# log of p(x)
p = gmm(test_coords, as_log=False)
N_s = 1000
# draw samples from GMM
samples = gmm.draw(N_s)
# draw sample from the model with noise, background, and selection:
# if you want to get the missing sample, set invert_sel=True.
# N_orig is the estimated number of samples prior to selection
obs_size = len(data)
samples, covar_samples, N_orig = pygmmis.draw(gmm, obs_size, sel_callback=cb,\
invert_sel=False, orig_size=None,\
covar_callback=covar_cb,background=bg)
```
For a complete example, have a look at [the test script](https://github.com/pmelchior/pygmmis/blob/master/tests/test.py). For requests and bug reports, please open an issue.
Raw data
{
"_id": null,
"home_page": "https://github.com/pmelchior/pygmmis",
"name": "pygmmis",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "",
"author": "Peter Melchior",
"author_email": "peter.m.melchior@gmail.com",
"download_url": "https://files.pythonhosted.org/packages/37/a0/6da1176287fcd36a48113db359a9a689d89a4f24634f1ee7b87e64f9a137/pygmmis-1.2.3.tar.gz",
"platform": null,
"description": "[](https://pypi.python.org/pypi/pygmmis/)\n[](https://github.com/pmelchior/pygmmis/blob/master/LICENSE.md)\n[](https://doi.org/10.1016/j.ascom.2018.09.013)\n[](http://arxiv.org/abs/1611.05806)\n\n# pyGMMis\n\nNeed a simple and powerful Gaussian-mixture code in pure python? It can be as easy as this:\n\n```python\nimport pygmmis\ngmm = pygmmis.GMM(K=K, D=D) # K components, D dimensions\nlogL, U = pygmmis.fit(gmm, data) # logL = log-likelihood, U = association of data to components\n```\nHowever, **pyGMMis** has a few extra tricks up its sleeve.\n\n* It can account for independent multivariate normal measurement errors for each of the observed samples, and then recovers an estimate of the error-free distribution. This technique is known as \"Extreme Deconvolution\" by Bovy, Hogg & Roweis (2011).\n* It works with missing data (features) by setting the respective elements of the covariance matrix to a vary large value, thus effectively setting the weights of the missing feature to 0.\n* It can deal with gaps (aka \"truncated data\") and variable sample completeness as long as\n * you know the incompleteness over the entire feature space,\n * and the incompleteness does not depend on the sample density (missing at random).\n* It can incorporate a \"background\" distribution (implemented is a uniform one) and separate signal from background, with the former being fit by the GMM.\n* It keeps track of which components need to be evaluated in which regions of the feature space, thereby substantially increasing the performance for fragmented data.\n\nIf you want more context and details on those capabilities, have a look at this [blog post](http://pmelchior.net/blog/gaussian-mixture-models-for-astronomy.html).\n\nUnder the hood, **pyGMMis** uses the Expectation-Maximization procedure. When dealing with sample incompleteness it generates its best guess of the unobserved samples on the fly given the current model fit to the observed samples.\n\n\n\nIn the example above, the true distribution is shown as contours in the left panel. We then draw 400 samples from it (red), add Gaussian noise to them (1,2,3 sigma contours shown in blue), and select only samples within the box but outside of the circle (blue).\n\nThe code is written in pure python (developed and tested in 2.7), parallelized with `multiprocessing`, and is capable of performing density estimation with millions of samples and thousands of model components on machines with sufficient memory.\n\nMore details are in the paper listed in the file `CITATION.cff`.\n\n\n\n## Installation and Prerequisites\n\nYou can either clone the repo and install by `python setup.py install` or get the latest release with\n\n```\npip install pygmmis\n```\n\nDependencies:\n\n* numpy\n* scipy\n* multiprocessing\n* parmap\n\n## How to run the code\n\n1. Create a GMM object with the desired component number K and data dimensionality D:\n ```gmm = pygmmis.GMM(K=K, D=D) ```\n\n3. Define a callback for the completeness function. When called with with `data` with shape `(N,D)` and returns the probability of each sample getting observed. Two simple examples:\n\n ```python\n def cutAtSix(coords):\n \t\"\"\"Selects all samples whose first coordinate is < 6\"\"\"\n return (coords[:,0] < 6)\n\n def selSlope(coords, rng=np.random):\n \"\"\"Selects probabilistically according to first coordinate x:\n Omega = 1 for x < 0\n = 1-x for x = 0 .. 1\n = 0 for x > 1\n \"\"\"\n return np.max(0, np.min(1, 1 - coords[:,0]))\n ```\n\n4. If the samples are noisy (i.e. they have positional uncertainties), you need to provide the covariance matrix of each data sample, or one for all in case of i.i.d. noise.\n\n4. If the samples are noisy *and* there completeness function isn't constant, you need to provide a callback function that returns an estimate of the covariance at arbitrary locations:\n\n ```python\n # example 1: simply using the same covariance for all samples\n dispersion = 1\n default_covar = np.eye(D) * dispersion**2\n covar_cb = lambda coords: default_covar\n \n # example: use the covariance of the nearest neighbor.\n def covar_tree_cb(coords, tree, covar):\n \"\"\"Return the covariance of the nearest neighbor of coords in data.\"\"\"\n dist, ind = tree.query(coords, k=1)\n return covar[ind.flatten()]\n \n from sklearn.neighbors import KDTree\n tree = KDTree(data, leaf_size=100)\n \n from functools import partial\n covar_cb = partial(covar_tree_cb, tree=tree, covar=covar)\n ```\n\n5. If there is a uniform background signal, you need to define it. Because a uniform distribution is normalizable only if its support is finite, you need to decide on the footprint over which the background model is present, e.g.:\n\n ```python\n footprint = data.min(axis=0), data.max(axis=0)\n amp = 0.3\n bg = pygmmis.Background(footprint, amp=amp)\n \n # fine tuning, if desired\n bg.amp_min = 0.1\n bg.amp_max = 0.5\n bg.adjust_amp = False # freezes bg.amp at current value\n ```\n\n6. Select an initialization method. This tells the GMM what initial parameters is should assume. The options are `'minmax','random','kmeans','none'`. See the respective functions for details:\n\n * `pygmmis.initFromDataMinMax()`\n * `pygmmis.initFromDataAtRandom()`\n * `pygmmis.initFromKMeans()`\n\n For difficult situations, or if you are not happy with the convergence, you may want to experiment with your own initialization. All you have to do is set `gmm.amp`, `gmm.mean`, and `gmm.covar` to desired values and use `init_method='none'`.\n\n7. Decide to freeze out any components. This makes sense if you *know* some of the parameters of the components. You can freeze amplitude, mean, or covariance of any component by listing them in a dictionary, e.g:\n\n ```python\n frozen={\"amp\": [1,2], \"mean\": [], \"covar\": [1]}\n ```\n\n This freezes the amplitudes of component 1 and 2 (NOTE: Counting starts at 0), and the covariance of 1.\n\n8. Run the fitter:\n\n ```python\n w = 0.1 # minimum covariance regularization, same units as data\n cutoff = 5 # segment the data set into neighborhood within 5 sigma around components\n tol = 1e-3 # tolerance on logL to terminate EM\n \n # define RNG for deterministic behavior\n from numpy.random import RandomState\n seed = 42\n rng = RandomState(seed)\n \n # run EM\n logL, U = pygmmis.fit(gmm, data, init_method='random',\\\n sel_callback=cb, covar_callback=covar_cb, w=w, cutoff=cutoff,\\\n background=bg, tol=tol, frozen=frozen, rng=rng)\n ```\n\n This runs the EM procedure until tolerance is reached and returns the final mean log-likelihood of all samples, and the neighborhood of each component (indices of data samples that are within cutoff of a GMM component).\n\n9. Evaluate the model:\n\n ```python\n # log of p(x)\n p = gmm(test_coords, as_log=False)\n N_s = 1000\n # draw samples from GMM\n samples = gmm.draw(N_s)\n \n # draw sample from the model with noise, background, and selection:\n # if you want to get the missing sample, set invert_sel=True.\n # N_orig is the estimated number of samples prior to selection\n obs_size = len(data)\n samples, covar_samples, N_orig = pygmmis.draw(gmm, obs_size, sel_callback=cb,\\\n invert_sel=False, orig_size=None,\\\n covar_callback=covar_cb,background=bg)\n ```\n\n\n\nFor a complete example, have a look at [the test script](https://github.com/pmelchior/pygmmis/blob/master/tests/test.py). For requests and bug reports, please open an issue.\n\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Gaussian mixture model for incomplete, truncated, and noisy data",
"version": "1.2.3",
"split_keywords": [],
"urls": [
{
"comment_text": "",
"digests": {
"md5": "78bf0a33561dc6059a12258dfe4c0958",
"sha256": "16484d0e4a7562dd72a1fc8b1e1b7335cba05489570bdbeb5e7cbfc80cca90f6"
},
"downloads": -1,
"filename": "pygmmis-1.2.3-py2.py3-none-any.whl",
"has_sig": false,
"md5_digest": "78bf0a33561dc6059a12258dfe4c0958",
"packagetype": "bdist_wheel",
"python_version": "py2.py3",
"requires_python": null,
"size": 22395,
"upload_time": "2022-07-21T02:41:56",
"upload_time_iso_8601": "2022-07-21T02:41:56.323284Z",
"url": "https://files.pythonhosted.org/packages/91/89/0d05f54d814eb025d422023a5972420672db1dc75048c05bb79cca8914fc/pygmmis-1.2.3-py2.py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"md5": "8211b2b88bbde4176d6d8614d2546b2f",
"sha256": "3bc0052a2b4c1ffc35ec66d0da6a5fcfdf0b845efc70029ff11321aa32e76203"
},
"downloads": -1,
"filename": "pygmmis-1.2.3.tar.gz",
"has_sig": false,
"md5_digest": "8211b2b88bbde4176d6d8614d2546b2f",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 22250,
"upload_time": "2022-07-21T02:41:58",
"upload_time_iso_8601": "2022-07-21T02:41:58.183765Z",
"url": "https://files.pythonhosted.org/packages/37/a0/6da1176287fcd36a48113db359a9a689d89a4f24634f1ee7b87e64f9a137/pygmmis-1.2.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2022-07-21 02:41:58",
"github": true,
"gitlab": false,
"bitbucket": false,
"github_user": "pmelchior",
"github_project": "pygmmis",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "pygmmis"
}
