# Mixture fitting library
```pymixfit``` is a Python mixture fitting library. This library tries
to fit a variety of candidate functions to presented measurement data
to extract the different components that make up the sampled data
points. This may be used to identify different mechanisms that contribute
to formation of data.
This library is based on fitting capabilities of [lmfit](https://lmfit.github.io//lmfit-py/).
The whole idea of mixture fitting and fitting sums using least squares
is built on the ideas of
* [Fitting mixtures of statistical distributions](https://arvix.org/abs/1901.06708)
* [Bundle adjustment methods in engineering photogrammetry](https://doi.org/10.1111/j.1477-9730.1980.tb00020.x)
## Installation
```
pip install pymixfit-tspspi
```
## Currently implemented model functions
* Constant (```mixfitfunctions.constant.MixfitFunctionConstantFactory```)
* $f(x) = \text{offset}$
* Linear (```mixfitfunctions.linear.MixfitFunctionLinearFactory```)
* $f(x) = \text{slope} * x + \text{intercept}$
* Gaussian (```mixfitfunctions.gaussian.MixfitFunctionGaussianFactory```)
* $f(x) = \text{amp} * \frac{1}{\sqrt{2 \pi}} e^{- \frac{1}{2} \left(\frac{x-\mu}{\sigma}\right)^2} + \text{offset}$
* Differential Gaussian (```mixfitfunctions.differentialgaussian.MixfitFunctionDifferentialGaussian```)
* Cauchy / Lorentz (```mixfitfunctions.cauchy.MixfitFunctionCauchyFactory```)
* Differential Cauchy / Lorentz (```mixfitfunctions.cauchy.MixfitFunctionDifferentialCauchyFactory```)
By default all functions are used as candidate functions by the mixture fitter
## Usage
To use the mixture fitter simply instantiate the ```Mixfit``` class and
perform a ```fit``` function. It's a good idea to apply one of the abort
conditions, else the fitter only aborts when it reaches the point of no
further improvements. Possible abort conditions are:
* ```stopError``` is the improvement of the $\chi^2$. As soon as the
fit quality of the fit goes below the threashold the process is aborted
* ```maxIterations``` limits the number of components that are fit
One may supply a list of allowed functions as well as their limits using
the ```allowed``` argument:
## Example
For more advanced examples take a look at the ```examples``` directory.
### Fitting arbitrary models into our data
```
import numpy as np
import matplotlib.pyplot as plt
from mixfit.mixfit import Mixfit
data = np.load("examplefile.npz")
# Just a way to get the sample data
# This data includes different runs capturing
# in-phase and quadrature channels during
# frequency sweeps. The x axis is the frequencies,
# the fitted data is the mean of all runs
x = data["f_RF"]
I = data["sigI"].mean(1)
# Now create the mixture fitter for _all_
# models
mf = Mixfit(
maxIterations = 4,
stopError = 0.05
)
# And execute
resI = mf.fit(x, I)
# Plot
fig, ax = plt.subplots(1, 2, figsize=(6.4*2, 4.8))
ax[0].plot(x*2, I) # We plot raw data
ax[0].plot(x*2, resI(x)) # and our fit result
ax[0].grid()
ax[1].plot(resI._chis)
ax[1].grid()
plt.show()
```
Running this code yields the following decomposition:
```
DiffGaussian(amp=6.710041629819328+-4.470775516222519, mu=175.76446887225788+-0.1454700003529027, sigma=1.4076661399565236+-0.3343414565050647, offset=-23320382.44684337+-112828498842278.73)
DiffGaussian(amp=2.774258665757228+-1.4958867327164893, mu=179.76706853968935+-0.4050248108850253, sigma=1.2976513505933047+-0.3279503072202676, offset=17745816.52197658+-175430109563261.66)
Cauchy(amp=-2.7037880990536274+-3.5613251200337626, x0=179.17057600546437+-0.08406908277066111, gamma=0.6377217109551133+-0.3774710838448468, offset=5574563.378103231+-166637348925010.78)
DiffCauchy(amp=0.01760683178016082+-0.03633273296203317, x0=175.6555822822984+-0.028894691836587848, gamma=0.06706371066814047+-0.12080936119483864, offset=0.002912072266518264+-4552116.210404506)
```
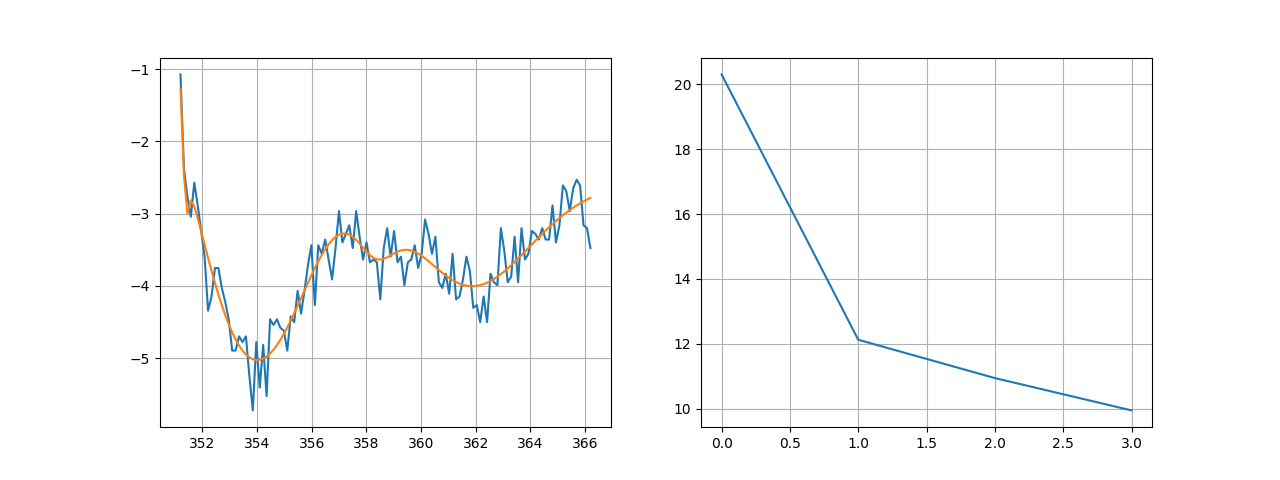
Raw data
{
"_id": null,
"home_page": "https://github.com/tspspi/pymixfit",
"name": "pymixfit-tspspi",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.6",
"maintainer_email": null,
"keywords": null,
"author": "Thomas Spielauer",
"author_email": "pypipackages01@tspi.at",
"download_url": "https://files.pythonhosted.org/packages/23/a0/e3a6f5b3691cfc9c5aedf6f11cf9e806c40966a7250216a684ea62fdf90c/pymixfit_tspspi-0.0.1.tar.gz",
"platform": null,
"description": "# Mixture fitting library\n\n```pymixfit``` is a Python mixture fitting library. This library tries\nto fit a variety of candidate functions to presented measurement data\nto extract the different components that make up the sampled data\npoints. This may be used to identify different mechanisms that contribute\nto formation of data.\n\nThis library is based on fitting capabilities of [lmfit](https://lmfit.github.io//lmfit-py/).\n\nThe whole idea of mixture fitting and fitting sums using least squares\nis built on the ideas of\n\n* [Fitting mixtures of statistical distributions](https://arvix.org/abs/1901.06708)\n* [Bundle adjustment methods in engineering photogrammetry](https://doi.org/10.1111/j.1477-9730.1980.tb00020.x)\n\n## Installation\n\n```\npip install pymixfit-tspspi\n```\n\n## Currently implemented model functions\n\n* Constant (```mixfitfunctions.constant.MixfitFunctionConstantFactory```)\n * $f(x) = \\text{offset}$\n* Linear (```mixfitfunctions.linear.MixfitFunctionLinearFactory```)\n * $f(x) = \\text{slope} * x + \\text{intercept}$\n* Gaussian (```mixfitfunctions.gaussian.MixfitFunctionGaussianFactory```)\n * $f(x) = \\text{amp} * \\frac{1}{\\sqrt{2 \\pi}} e^{- \\frac{1}{2} \\left(\\frac{x-\\mu}{\\sigma}\\right)^2} + \\text{offset}$\n* Differential Gaussian (```mixfitfunctions.differentialgaussian.MixfitFunctionDifferentialGaussian```)\n* Cauchy / Lorentz (```mixfitfunctions.cauchy.MixfitFunctionCauchyFactory```)\n* Differential Cauchy / Lorentz (```mixfitfunctions.cauchy.MixfitFunctionDifferentialCauchyFactory```)\n\nBy default all functions are used as candidate functions by the mixture fitter\n\n## Usage\n\nTo use the mixture fitter simply instantiate the ```Mixfit``` class and\nperform a ```fit``` function. It's a good idea to apply one of the abort\nconditions, else the fitter only aborts when it reaches the point of no\nfurther improvements. Possible abort conditions are:\n\n* ```stopError``` is the improvement of the $\\chi^2$. As soon as the\n fit quality of the fit goes below the threashold the process is aborted\n* ```maxIterations``` limits the number of components that are fit\n\nOne may supply a list of allowed functions as well as their limits using\nthe ```allowed``` argument:\n\n## Example\n\nFor more advanced examples take a look at the ```examples``` directory.\n\n### Fitting arbitrary models into our data\n\n```\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom mixfit.mixfit import Mixfit\n\ndata = np.load(\"examplefile.npz\")\n\n# Just a way to get the sample data\n# This data includes different runs capturing\n# in-phase and quadrature channels during\n# frequency sweeps. The x axis is the frequencies,\n# the fitted data is the mean of all runs\nx = data[\"f_RF\"]\nI = data[\"sigI\"].mean(1)\n\n# Now create the mixture fitter for _all_\n# models\nmf = Mixfit(\n\tmaxIterations = 4,\n\tstopError = 0.05\n)\n\n# And execute\nresI = mf.fit(x, I)\n\n# Plot\nfig, ax = plt.subplots(1, 2, figsize=(6.4*2, 4.8))\nax[0].plot(x*2, I) # We plot raw data\nax[0].plot(x*2, resI(x)) # and our fit result\nax[0].grid()\n\nax[1].plot(resI._chis)\nax[1].grid()\n\nplt.show()\n```\n\nRunning this code yields the following decomposition:\n\n```\nDiffGaussian(amp=6.710041629819328+-4.470775516222519, mu=175.76446887225788+-0.1454700003529027, sigma=1.4076661399565236+-0.3343414565050647, offset=-23320382.44684337+-112828498842278.73)\nDiffGaussian(amp=2.774258665757228+-1.4958867327164893, mu=179.76706853968935+-0.4050248108850253, sigma=1.2976513505933047+-0.3279503072202676, offset=17745816.52197658+-175430109563261.66)\nCauchy(amp=-2.7037880990536274+-3.5613251200337626, x0=179.17057600546437+-0.08406908277066111, gamma=0.6377217109551133+-0.3774710838448468, offset=5574563.378103231+-166637348925010.78)\nDiffCauchy(amp=0.01760683178016082+-0.03633273296203317, x0=175.6555822822984+-0.028894691836587848, gamma=0.06706371066814047+-0.12080936119483864, offset=0.002912072266518264+-4552116.210404506)\n```\n\n\n\n",
"bugtrack_url": null,
"license": null,
"summary": "Mixture fitting",
"version": "0.0.1",
"project_urls": {
"Homepage": "https://github.com/tspspi/pymixfit"
},
"split_keywords": [],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "38d1d6f3692d8ae95c23be550f2bcd65baf7d510a5d738b297d8781a60fbec56",
"md5": "69c7ec1796290e9ad3d956950ed1252d",
"sha256": "37d18c4f1ca9ac7a4f712c23457a089a881d561b22b0cbca31feb434dc421c71"
},
"downloads": -1,
"filename": "pymixfit_tspspi-0.0.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "69c7ec1796290e9ad3d956950ed1252d",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 14255,
"upload_time": "2024-05-19T10:36:26",
"upload_time_iso_8601": "2024-05-19T10:36:26.915000Z",
"url": "https://files.pythonhosted.org/packages/38/d1/d6f3692d8ae95c23be550f2bcd65baf7d510a5d738b297d8781a60fbec56/pymixfit_tspspi-0.0.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "23a0e3a6f5b3691cfc9c5aedf6f11cf9e806c40966a7250216a684ea62fdf90c",
"md5": "3f6ebb5d584d8e28fd9d95f74dbd10fc",
"sha256": "b7b388dd203140177703eaafa2c921147e1f5e17aff489fd26e3749d586cb874"
},
"downloads": -1,
"filename": "pymixfit_tspspi-0.0.1.tar.gz",
"has_sig": false,
"md5_digest": "3f6ebb5d584d8e28fd9d95f74dbd10fc",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 11817,
"upload_time": "2024-05-19T10:36:29",
"upload_time_iso_8601": "2024-05-19T10:36:29.379460Z",
"url": "https://files.pythonhosted.org/packages/23/a0/e3a6f5b3691cfc9c5aedf6f11cf9e806c40966a7250216a684ea62fdf90c/pymixfit_tspspi-0.0.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-05-19 10:36:29",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "tspspi",
"github_project": "pymixfit",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "pymixfit-tspspi"
}
