| Name | qcio JSON |
| Version |
0.16.0
 JSON
JSON |
| download |
| home_page | None |
| Summary | Beautiful and user friendly data structures for quantum chemistry. |
| upload_time | 2025-10-09 18:38:54 |
| maintainer | None |
| docs_url | None |
| author | None |
| requires_python | >=3.10 |
| license | The MIT License (MIT)
Copyright (c) 2021 Colton Hicks
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in
all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN
THE SOFTWARE. |
| keywords |
cheminformatics
data-structures
quantum-chemistry
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
# Quantum Chemistry I/O
[](https://pypi.python.org/pypi/qcio)
[](https://pypi.python.org/pypi/qcio)
[](https://pypi.python.org/pypi/qcio)
[](https://github.com/coltonbh/qcio/actions)
[](https://github.com/coltonbh/qcio/actions)
Elegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations.
`qcio` works in harmony with a suite of other quantum chemistry tools for fast, structured, and interoperable quantum chemistry.
## The QC Suite of Programs
- [qcconst](https://github.com/coltonbh/qcconst) - NIST/CODATA2022 core physical constants, conversion factors, and a periodic table with clear source information for every value.
- [qcio](https://github.com/coltonbh/qcio) - Elegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations. [Documentation](https://qcio.coltonhicks.com)
- [qccodec](https://github.com/coltonbh/qccodec) - A translation layer between quantum chemistry program inputs and outputs and structured `qcio` objects.
- [qcop](https://github.com/coltonbh/qcop) - A package for operating quantum chemistry programs using `qcio` standardized data structures. Compatible with `TeraChem`, `psi4`, `QChem`, `NWChem`, `ORCA`, `Molpro`, `geomeTRIC` and many more.
- [BigChem](https://github.com/mtzgroup/bigchem) - A distributed application for running quantum chemistry calculations at scale across clusters of computers or the cloud. Bring multi-node scaling to your favorite quantum chemistry program.
- `ChemCloud` - A [web application](https://github.com/mtzgroup/chemcloud-server) and associated [Python client](https://github.com/mtzgroup/chemcloud-client) for exposing a BigChem cluster securely over the internet.
## Installation
```bash
python -m pip install qcio
```
## Quickstart
`qcio` is built around a simple mental model: `Input` objects define quantum chemistry calculations, and a `Results` object defines the results.
All `qcio` objects can be serialized and saved to disk by calling `.save("filename.json")` and loaded from disk by calling `.open("filename.json")`. `qcio` supports `json`, `yaml`, and `toml` file formats. Binary data will be automatically base64 encoded and decoded when saving and loading.
### Input Objects
#### CalcInput - Core input object for a single QC calculation.
```python
from qcio import Structure, CalcInput
# xyz files or saved Structure objects can be opened from disk
caffeine = Structure.open("caffeine.xyz")
# Define the program input
prog_input = CalcInput(
structure=caffeine,
calctype="energy",
keywords={"purify": "no", "restricted": False},
model={"method": "hf", "basis": "sto-3g"},
extras={"comment": "This is a comment"}, # Anything extra not in the schema
)
# Binary or other files used as input can be added
prog_input.add_file("wfn.dat")
prog_input.keywords["initial_guess"] = "wfn.dat"
# Save the input to disk in json, yaml, or toml format
prog_input.save("input.json")
# Open the input from disk
prog_input = CalcInput.open("input.json")
```
`Structure` objects can be opened from and saved as xyz files or saved to disk as `.json`, `.yaml`, or `.toml` formats by changing the extension of the file. For `.xyz` files precision can be controlled by passing the `precision` argument to the `save` method.
```python
caffeine = Structure.open("caffeine.xyz")
caffeine.save("caffeine2.json", precision=6)
caffeine.save("caffeine.toml")
```
#### CompositeCalcInput - Input object for a workflow that uses multiple QC calculations.
`CompositeCalcInput` objects can be used to power workflows that require multiple calculations. For example, a geometry optimization workflow might use `geomeTRIC` to power the optimization and use `terachem` to compute the energies and gradients.
```python
from qcio import Structure, CompositeCalcInput
# xyz files or saved Structure objects can be opened from disk
caffeine = Structure.open("caffeine.xyz")
# Define the program input
prog_input = CompositeCalcInput(
structure=caffeine,
calctype="optimization",
keywords={"maxiter": 250},
subprogram="terachem",
subprogram_args = {
"model": {"method": "hf", "basis": "sto-3g"},
"keywords": {"purify": "no", "restricted": False},
},
extras={"comment": "This is a comment"}, # Anything extra not in the schema
)
```
#### FileInput - Input object for a calculation using native file formats.
`qcio` also supports the native file formats of each QC program with a `FileInput` object. Assume you have a directory like this with your input files for `psi4`:
```
psi4/
input.dat
geometry.xyz
wfn.dat
```
You can collect these native files and any associated command line arguments needed to specify a calculation into a `FileInput` object like this:
```python
from qcio import FileInput
psi4_input = FileInput.from_directory("psi4")
# All input files will be loaded into the `files` attribute
psi4_input.files
# {'input.dat': '...', 'geometry.xyz': '...', 'wfn.dat': '...'}
# Add psi4 command line args to the input
psi4_input.cmdline_args.extend(["-n", "4"])
# Files can be dumped to a directory for a calculation
psi4_input.save_files("psi4")
```
#### Modifying Input Objects
Objects are immutable by default so if you want to modify an object cast it to a dictionary, make the desired modification, and then instantiate a new object. This prevents accidentally modifying objects that may already be referenced in other calculations--perhaps as `.input_data` on a `Results` object.
```python
# Cast to a dictionary and modify
new_input_dict = prog_input.model_dumps()
new_input_dict["model"]["method"] = "b3lyp"
# Instantiate a new object
new_prog_input = CalcInput(**new_input_dict)
```
### Results
Calculation values are stored in a `Results` object. `Results` may contain parsed data, files, logs, and additional details of the calculation. A `Results` object has the following attributes:
```python
output.input_data # Input data used for the calculation
output.success # Whether the calculation succeeded or failed
output.data # All structured data from the calculation
output.data.files # Any files returned by the calculation
output.logs # Logs from the calculation
output.plogs # Shortcut to print the logs in human readable format
output.provenance # Provenance information about the calculation
output.extras # Any extra information not in the schema
```
The `.data` attribute on a `Results` is polymorphic and may be either `Files`, `SinglePointData`, `OptimizationData`, `ConformerSearchData`, or any other `DataType` depending on the calculation requested. Available attributes for each data type can be found by calling `dir()` on the object.
```python
dir(results.data)
```
Results can be saved to disk in json, yaml, or toml format by calling `.save("filename.{json/yaml/toml}")` and loaded from disk by calling `.open("filename.{json/yaml/toml}")`.
## ✨ Visualization ✨
Visualize all your results with a single line of code!
First install the visualization module:
```sh
python -m pip install qcio[view]
```
or if your shell requires `''` around arguments with brackets:
```sh
python -m pip install 'qcio[view]'
```
Then in a Jupyter notebook import the `qcio` view module and call `view.view(...)` passing it one or any number of `qcio` objects you want to visualizing including `Structure` objects or any `Results` object. You may also pass an array of `titles` and/or `subtitles` to add additional information to the molecular structure display. If no titles are passed `qcio` with look for `Structure` identifiers such as a name or SMILES to label the `Structure`.
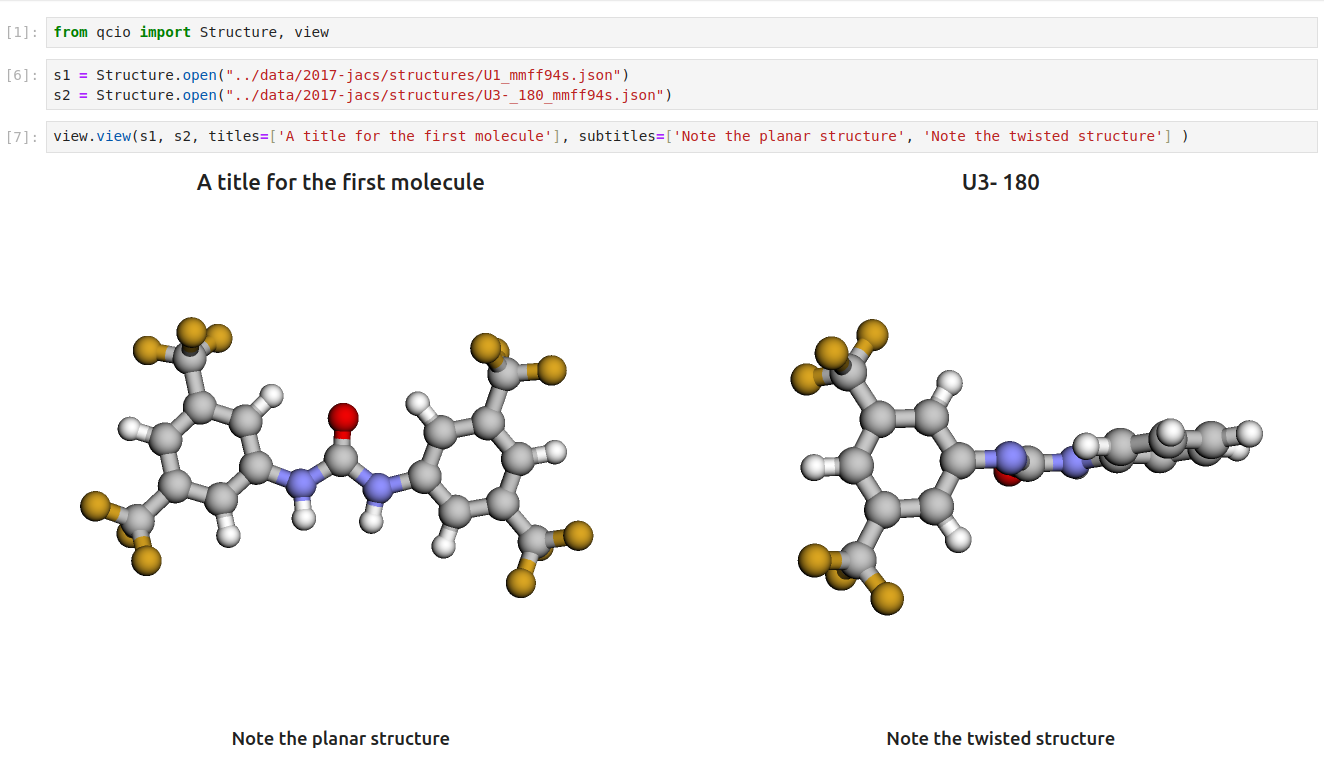
Seamless visualizations for `Results` objects make results analysis easy!

Single point calculations display their results in a table.

If you want to use the HTML generated by the viewer to build your own dashboards use the functions inside of `qcio.view.py` that begin with the word `generate_` to create HTML you can insert into any dashboard.
## Development
To get started with all development dependencies:
```sh
uv sync --all-groups
```
## Support
If you have any issues with `qcio` or would like to request a feature, please open an [issue](https://github.com/coltonbh/qcio/issues).
Raw data
{
"_id": null,
"home_page": null,
"name": "qcio",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": null,
"keywords": "cheminformatics, data-structures, quantum-chemistry",
"author": null,
"author_email": "Colton Hicks <github@coltonhicks.com>",
"download_url": "https://files.pythonhosted.org/packages/ad/93/870c0e5ee6f4879e6942094d563ba0ad61fe5ccb0af24b6d2049534c1e87/qcio-0.16.0.tar.gz",
"platform": null,
"description": "# Quantum Chemistry I/O\n\n[](https://pypi.python.org/pypi/qcio)\n[](https://pypi.python.org/pypi/qcio)\n[](https://pypi.python.org/pypi/qcio)\n[](https://github.com/coltonbh/qcio/actions)\n[](https://github.com/coltonbh/qcio/actions)\n\nElegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations.\n\n`qcio` works in harmony with a suite of other quantum chemistry tools for fast, structured, and interoperable quantum chemistry.\n\n## The QC Suite of Programs\n\n- [qcconst](https://github.com/coltonbh/qcconst) - NIST/CODATA2022 core physical constants, conversion factors, and a periodic table with clear source information for every value.\n- [qcio](https://github.com/coltonbh/qcio) - Elegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations. [Documentation](https://qcio.coltonhicks.com)\n- [qccodec](https://github.com/coltonbh/qccodec) - A translation layer between quantum chemistry program inputs and outputs and structured `qcio` objects.\n- [qcop](https://github.com/coltonbh/qcop) - A package for operating quantum chemistry programs using `qcio` standardized data structures. Compatible with `TeraChem`, `psi4`, `QChem`, `NWChem`, `ORCA`, `Molpro`, `geomeTRIC` and many more.\n- [BigChem](https://github.com/mtzgroup/bigchem) - A distributed application for running quantum chemistry calculations at scale across clusters of computers or the cloud. Bring multi-node scaling to your favorite quantum chemistry program.\n- `ChemCloud` - A [web application](https://github.com/mtzgroup/chemcloud-server) and associated [Python client](https://github.com/mtzgroup/chemcloud-client) for exposing a BigChem cluster securely over the internet.\n\n## Installation\n\n```bash\npython -m pip install qcio\n```\n\n## Quickstart\n\n`qcio` is built around a simple mental model: `Input` objects define quantum chemistry calculations, and a `Results` object defines the results.\n\nAll `qcio` objects can be serialized and saved to disk by calling `.save(\"filename.json\")` and loaded from disk by calling `.open(\"filename.json\")`. `qcio` supports `json`, `yaml`, and `toml` file formats. Binary data will be automatically base64 encoded and decoded when saving and loading.\n\n### Input Objects\n\n#### CalcInput - Core input object for a single QC calculation.\n\n```python\nfrom qcio import Structure, CalcInput\n# xyz files or saved Structure objects can be opened from disk\ncaffeine = Structure.open(\"caffeine.xyz\")\n# Define the program input\nprog_input = CalcInput(\n structure=caffeine,\n calctype=\"energy\",\n keywords={\"purify\": \"no\", \"restricted\": False},\n model={\"method\": \"hf\", \"basis\": \"sto-3g\"},\n extras={\"comment\": \"This is a comment\"}, # Anything extra not in the schema\n)\n# Binary or other files used as input can be added\nprog_input.add_file(\"wfn.dat\")\nprog_input.keywords[\"initial_guess\"] = \"wfn.dat\"\n\n# Save the input to disk in json, yaml, or toml format\nprog_input.save(\"input.json\")\n\n# Open the input from disk\nprog_input = CalcInput.open(\"input.json\")\n```\n\n`Structure` objects can be opened from and saved as xyz files or saved to disk as `.json`, `.yaml`, or `.toml` formats by changing the extension of the file. For `.xyz` files precision can be controlled by passing the `precision` argument to the `save` method.\n\n```python\ncaffeine = Structure.open(\"caffeine.xyz\")\ncaffeine.save(\"caffeine2.json\", precision=6)\ncaffeine.save(\"caffeine.toml\")\n```\n\n#### CompositeCalcInput - Input object for a workflow that uses multiple QC calculations.\n\n`CompositeCalcInput` objects can be used to power workflows that require multiple calculations. For example, a geometry optimization workflow might use `geomeTRIC` to power the optimization and use `terachem` to compute the energies and gradients.\n\n```python\nfrom qcio import Structure, CompositeCalcInput\n# xyz files or saved Structure objects can be opened from disk\ncaffeine = Structure.open(\"caffeine.xyz\")\n# Define the program input\nprog_input = CompositeCalcInput(\n structure=caffeine,\n calctype=\"optimization\",\n keywords={\"maxiter\": 250},\n subprogram=\"terachem\",\n subprogram_args = {\n \"model\": {\"method\": \"hf\", \"basis\": \"sto-3g\"},\n \"keywords\": {\"purify\": \"no\", \"restricted\": False},\n },\n extras={\"comment\": \"This is a comment\"}, # Anything extra not in the schema\n)\n```\n\n#### FileInput - Input object for a calculation using native file formats.\n\n`qcio` also supports the native file formats of each QC program with a `FileInput` object. Assume you have a directory like this with your input files for `psi4`:\n\n```\npsi4/\n input.dat\n geometry.xyz\n wfn.dat\n```\n\nYou can collect these native files and any associated command line arguments needed to specify a calculation into a `FileInput` object like this:\n\n```python\nfrom qcio import FileInput\npsi4_input = FileInput.from_directory(\"psi4\")\n\n# All input files will be loaded into the `files` attribute\npsi4_input.files\n# {'input.dat': '...', 'geometry.xyz': '...', 'wfn.dat': '...'}\n\n# Add psi4 command line args to the input\npsi4_input.cmdline_args.extend([\"-n\", \"4\"])\n\n# Files can be dumped to a directory for a calculation\npsi4_input.save_files(\"psi4\")\n```\n\n#### Modifying Input Objects\n\nObjects are immutable by default so if you want to modify an object cast it to a dictionary, make the desired modification, and then instantiate a new object. This prevents accidentally modifying objects that may already be referenced in other calculations--perhaps as `.input_data` on a `Results` object.\n\n```python\n# Cast to a dictionary and modify\nnew_input_dict = prog_input.model_dumps()\nnew_input_dict[\"model\"][\"method\"] = \"b3lyp\"\n# Instantiate a new object\nnew_prog_input = CalcInput(**new_input_dict)\n```\n\n### Results\n\nCalculation values are stored in a `Results` object. `Results` may contain parsed data, files, logs, and additional details of the calculation. A `Results` object has the following attributes:\n\n```python\noutput.input_data # Input data used for the calculation\noutput.success # Whether the calculation succeeded or failed\noutput.data # All structured data from the calculation\noutput.data.files # Any files returned by the calculation\noutput.logs # Logs from the calculation\noutput.plogs # Shortcut to print the logs in human readable format\noutput.provenance # Provenance information about the calculation\noutput.extras # Any extra information not in the schema\n```\n\nThe `.data` attribute on a `Results` is polymorphic and may be either `Files`, `SinglePointData`, `OptimizationData`, `ConformerSearchData`, or any other `DataType` depending on the calculation requested. Available attributes for each data type can be found by calling `dir()` on the object.\n\n```python\ndir(results.data)\n```\n\nResults can be saved to disk in json, yaml, or toml format by calling `.save(\"filename.{json/yaml/toml}\")` and loaded from disk by calling `.open(\"filename.{json/yaml/toml}\")`.\n\n## \u2728 Visualization \u2728\n\nVisualize all your results with a single line of code!\n\nFirst install the visualization module:\n\n```sh\npython -m pip install qcio[view]\n```\n\nor if your shell requires `''` around arguments with brackets:\n\n```sh\npython -m pip install 'qcio[view]'\n```\n\nThen in a Jupyter notebook import the `qcio` view module and call `view.view(...)` passing it one or any number of `qcio` objects you want to visualizing including `Structure` objects or any `Results` object. You may also pass an array of `titles` and/or `subtitles` to add additional information to the molecular structure display. If no titles are passed `qcio` with look for `Structure` identifiers such as a name or SMILES to label the `Structure`.\n\n\n\nSeamless visualizations for `Results` objects make results analysis easy!\n\n\n\nSingle point calculations display their results in a table.\n\n\n\nIf you want to use the HTML generated by the viewer to build your own dashboards use the functions inside of `qcio.view.py` that begin with the word `generate_` to create HTML you can insert into any dashboard.\n\n## Development\n\nTo get started with all development dependencies:\n\n```sh\nuv sync --all-groups\n```\n\n## Support\n\nIf you have any issues with `qcio` or would like to request a feature, please open an [issue](https://github.com/coltonbh/qcio/issues).\n",
"bugtrack_url": null,
"license": "The MIT License (MIT)\n \n Copyright (c) 2021 Colton Hicks\n \n Permission is hereby granted, free of charge, to any person obtaining a copy\n of this software and associated documentation files (the \"Software\"), to deal\n in the Software without restriction, including without limitation the rights\n to use, copy, modify, merge, publish, distribute, sublicense, and/or sell\n copies of the Software, and to permit persons to whom the Software is\n furnished to do so, subject to the following conditions:\n \n The above copyright notice and this permission notice shall be included in\n all copies or substantial portions of the Software.\n \n THE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\n IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\n FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\n AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\n LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\n OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN\n THE SOFTWARE.",
"summary": "Beautiful and user friendly data structures for quantum chemistry.",
"version": "0.16.0",
"project_urls": {
"Bug Tracker": "https://github.com/coltonbh/qcio/issues",
"Changelog": "https://github.com/coltonbh/qcio/blob/master/CHANGELOG.md",
"Documentation": "https://qcio.coltonhicks.com",
"Homepage": "https://github.com/coltonbh/qcio",
"Source": "https://github.com/coltonbh/qcio",
"repository": "https://github.com/coltonbh/qcio"
},
"split_keywords": [
"cheminformatics",
" data-structures",
" quantum-chemistry"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "b2d342d945e34abc54af2eb4454824dc63d29316b87cb58f72d4248fabff2278",
"md5": "5e830340be4089c9450d9a994b9a73f8",
"sha256": "d8fbd19d32a19ff2c3843b004cdc80b014ce2d8eff8714406fe4a9387143b6b7"
},
"downloads": -1,
"filename": "qcio-0.16.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "5e830340be4089c9450d9a994b9a73f8",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10",
"size": 36723,
"upload_time": "2025-10-09T18:38:52",
"upload_time_iso_8601": "2025-10-09T18:38:52.875814Z",
"url": "https://files.pythonhosted.org/packages/b2/d3/42d945e34abc54af2eb4454824dc63d29316b87cb58f72d4248fabff2278/qcio-0.16.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "ad93870c0e5ee6f4879e6942094d563ba0ad61fe5ccb0af24b6d2049534c1e87",
"md5": "804c1e0dd092011b7c2dda52411cbcaa",
"sha256": "5b803e9d831fb2280ac43c48f87b76e0c8150391c1d4d561f749f0a4f4c5e0cb"
},
"downloads": -1,
"filename": "qcio-0.16.0.tar.gz",
"has_sig": false,
"md5_digest": "804c1e0dd092011b7c2dda52411cbcaa",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 1473997,
"upload_time": "2025-10-09T18:38:54",
"upload_time_iso_8601": "2025-10-09T18:38:54.834922Z",
"url": "https://files.pythonhosted.org/packages/ad/93/870c0e5ee6f4879e6942094d563ba0ad61fe5ccb0af24b6d2049534c1e87/qcio-0.16.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-10-09 18:38:54",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "coltonbh",
"github_project": "qcio",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "qcio"
}
