| Name | qcop JSON |
| Version |
0.9.8
 JSON
JSON |
| download |
| home_page | None |
| Summary | A package for operating Quantum Chemistry programs using qcio standardized data structures. Compatible with TeraChem, psi4, QChem, NWChem, ORCA, Molpro, geomeTRIC and many more. |
| upload_time | 2025-03-01 00:58:40 |
| maintainer | None |
| docs_url | None |
| author | Colton Hicks |
| requires_python | <4.0,>=3.9 |
| license | MIT |
| keywords |
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
# Quantum Chemistry Operate
A package for operating Quantum Chemistry programs using [qcio](https://github.com/coltonbh/qcio) standardized data structures. Compatible with `TeraChem`, `psi4`, `QChem`, `NWChem`, `ORCA`, `Molpro`, `geomeTRIC` and many more.
[](https://pypi.python.org/pypi/qcop)
[](https://pypi.python.org/pypi/qcop)
[](https://pypi.python.org/pypi/qcop)
[](https://github.com/coltonbh/qcop/actions)
[](https://github.com/coltonbh/qcop/actions)
[](https://github.com/charliermarsh/ruff)
`qcop` works in harmony with a suite of other quantum chemistry tools for fast, structured, and interoperable quantum chemistry.
## The QC Suite of Programs
- [qcio](https://github.com/coltonbh/qcio) - Elegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations.
- [qcparse](https://github.com/coltonbh/qcparse) - A library for efficient parsing of quantum chemistry data into structured `qcio` objects.
- [qcop](https://github.com/coltonbh/qcop) - A package for operating quantum chemistry programs using `qcio` standardized data structures. Compatible with `TeraChem`, `psi4`, `QChem`, `NWChem`, `ORCA`, `Molpro`, `geomeTRIC`, and many more, featuring seamless Jupyter Notebook visualizations.
- [BigChem](https://github.com/mtzgroup/bigchem) - A distributed application for running quantum chemistry calculations at scale across clusters of computers or the cloud. Bring multi-node scaling to your favorite quantum chemistry program, featuring seamless Jupyter Notebook visualizations.
- `ChemCloud` - A [web application](https://github.com/mtzgroup/chemcloud-server) and associated [Python client](https://github.com/mtzgroup/chemcloud-client) for exposing a BigChem cluster securely over the internet, featuring seamless Jupyter Notebook visualizations.
## Installation
```sh
pip install qcop
```
## Quickstart
`qcop` uses the `qcio` data structures to drive quantum chemistry programs in a standardized way. This allows for a simple and consistent interface to a wide variety of quantum chemistry programs. See the [qcio](https://github.com/coltonbh/qcio) library for documentation on the input and output data structures.
The `compute` function is the main entry point for the library and is used to run a calculation.
```python
from qcio import Structure, ProgramInput
from qcop import compute
from qcop.exceptions import ExternalProgramError
# Create the Structure
h2o = Structure.open("h2o.xyz")
# Define the program input
prog_input = ProgramInput(
structure=h2o,
calctype="energy",
model={"method": "hf", "basis": "sto-3g"},
keywords={"purify": "no", "restricted": False},
)
# Run the calculation; will return ProgramOutput or raise an exception
try:
po = compute("terachem", prog_input, collect_files=True)
except ExternalProgramError as e:
# External QQ program failed in some way
po = e.program_output
po.input_data # Input data used by the QC program
po.success # Will be False
po.results # Any half-computed results before the failure
po.traceback # Stack trace from the calculation
po.ptraceback # Shortcut to print out the traceback in human readable format
po.stdout # Stdout log from the calculation
raise e
else:
# Calculation succeeded
po.input_data # Input data used by the QC program
po.success # Will be True
po.results # All structured results from the calculation
po.stdout # Stdout log from the calculation
po.pstdout # Shortcut to print out the stdout in human readable format
po.files # Any files returned by the calculation
po.provenance # Provenance information about the calculation
po.extras # Any extra information not in the schema
```
One may also call `compute(..., raise_exc=False)` to return a `ProgramOutput` object rather than raising an exception when a calculation fails. This may allow easier handling of failures in some cases.
```python
from qcio import Structure, ProgramInput
from qcop import compute
from qcop.exceptions import ExternalProgramError
# Create the Structure
h2o = Structure.open("h2o.xyz")
# Define the program input
prog_input = ProgramInput(
structure=h2o,
calctype="energy",
model={"method": "hf", "basis": "sto-3g"},
keywords={"purify": "no", "restricted": False},
)
# Run the calculation; will return a ProgramOutput objects
po = compute("terachem", prog_input, collect_files=True, raise_exc=False)
if not po.success:
# External QQ program failed in some way
po.input_data # Input data used by the QC program
po.success # Will be False
po.results # Any half-computed results before the failure
po.traceback # Stack trace from the calculation
po.ptraceback # Shortcut to print out the traceback in human readable format
po.stdout # Stdout log from the calculation
else:
# Calculation succeeded
po.input_data # Input data used by the QC program
po.success # Will be True
po.results # All structured results from the calculation
po.stdout # Stdout log from the calculation
po.pstdout # Shortcut to print out the stdout in human readable format
po.files # Any files returned by the calculation
po.provenance # Provenance information about the calculation
po.extras # Any extra information not in the schema
```
Alternatively, the `compute_args` function can be used to run a calculation with the input data structures passed in as arguments rather than as a single `ProgramInput` object.
```python
from qcio import Structure
from qcop import compute_args
# Create the Structure
h2o = Structure.open("h2o.xyz")
# Run the calculation
output = compute_args(
"terachem",
h2o,
calctype="energy",
model={"method": "hf", "basis": "sto-3g"},
keywords={"purify": "no", "restricted": False},
files={...},
collect_files=True
)
```
The behavior of `compute()` and `compute_args()` can be tuned by passing in keyword arguments like `collect_files` shown above. Keywords can modify which scratch directory location to use, whether to delete or keep the scratch files after a calculation completes, what files to collect from a calculation, whether to print the program stdout in real time as the program executes, and whether to propagate a wavefunction through a series of calculations. Keywords also include hooks for passing in update functions that can be called as a program executes in real time. See the [compute method docstring](https://github.com/coltonbh/qcop/blob/83868df51d241ffae3497981dfc3c72235319c6e/qcop/adapters/base.py#L57-L123) for more details.
See the [/examples](https://github.com/coltonbh/qcop/tree/master/examples) directory for more examples.
## ✨ Visualization ✨
Visualize all your results with a single line of code!
First install the visualization module:
```sh
pip install qcio[view]
```
or if your shell requires `''` around arguments with brackets:
```sh
pip install 'qcio[view]'
```
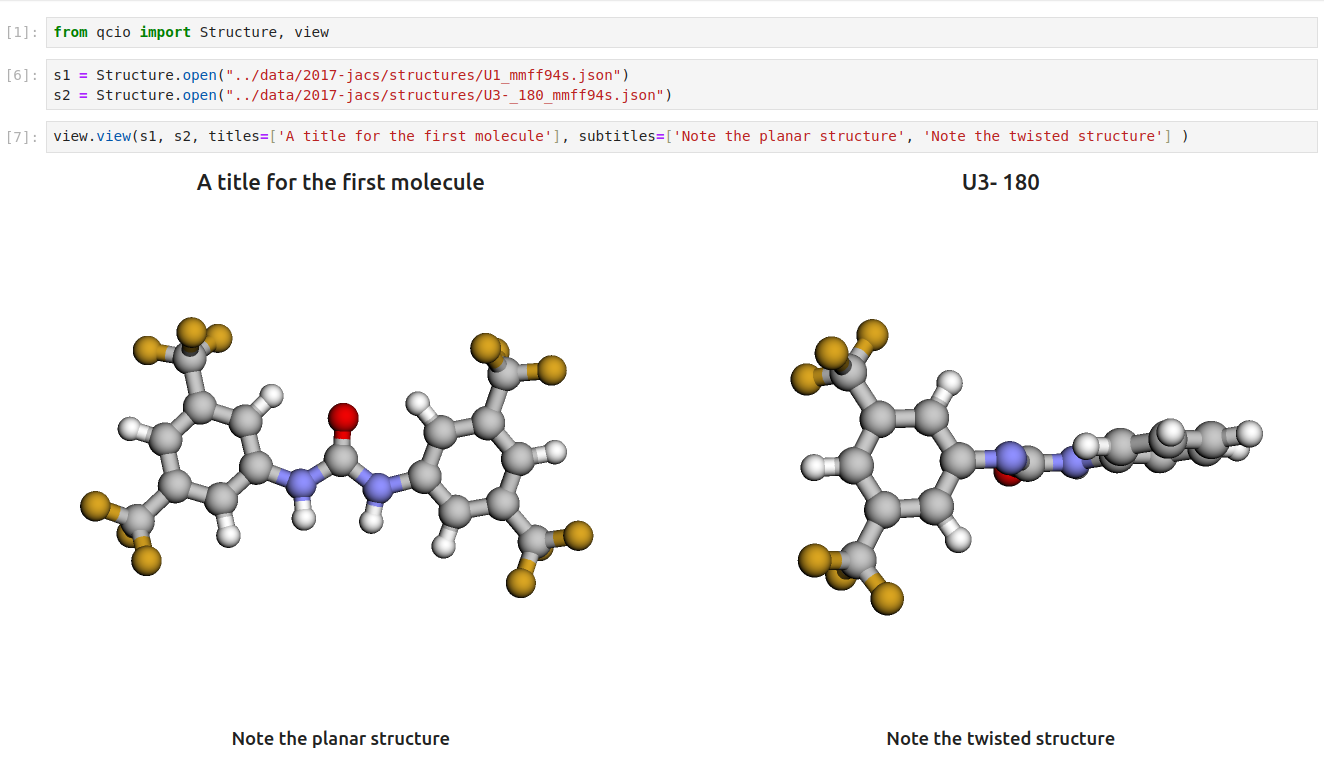
Then in a Jupyter notebook import the `qcio` view module and call `view.view(...)` passing it one or any number of `qcio` objects you want to visualizing including `Structure` objects or any `ProgramOutput` object. You may also pass an array of `titles` and/or `subtitles` to add additional information to the molecular structure display. If no titles are passed `qcio` with look for `Structure` identifiers such as a name or SMILES to label the `Structure`.
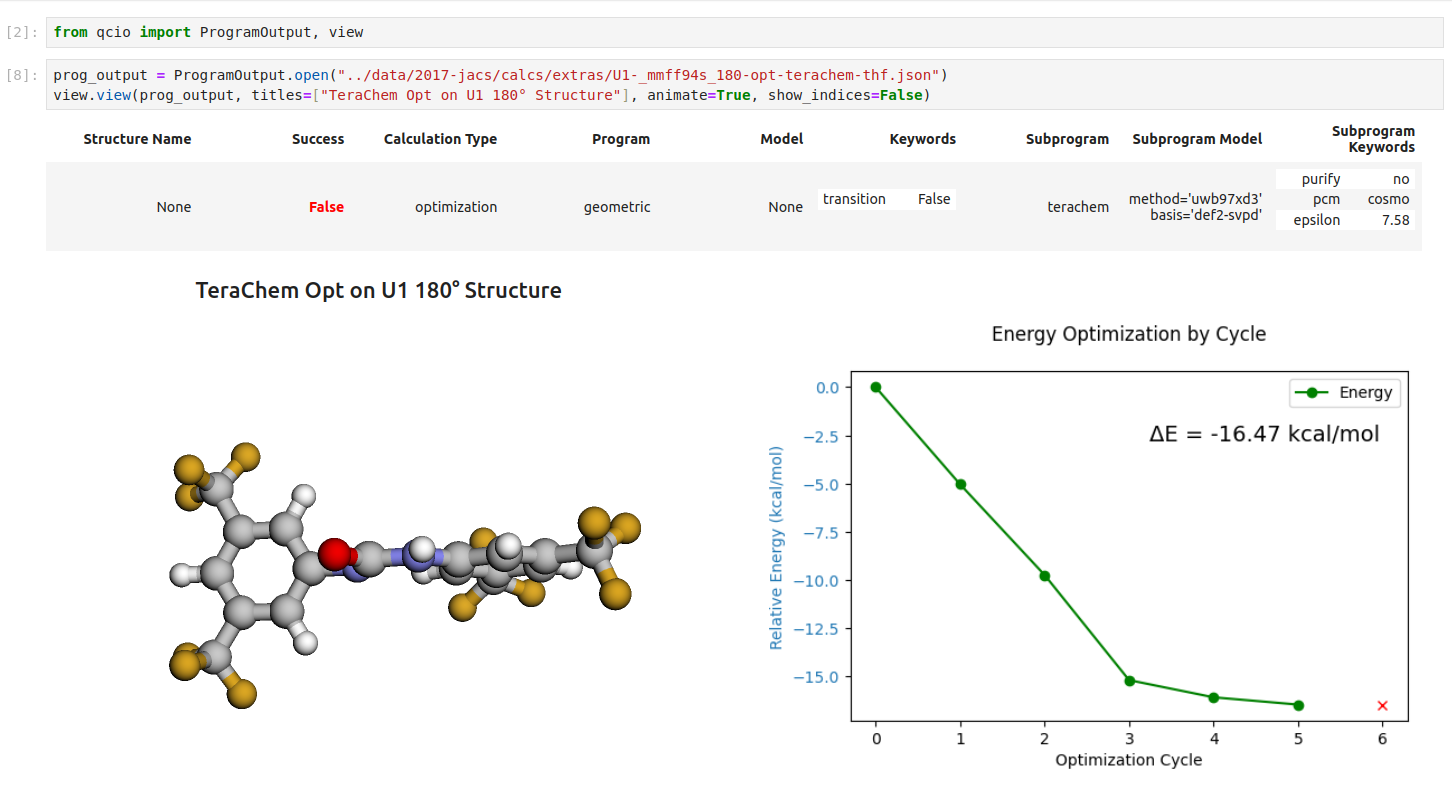
Seamless visualizations for `ProgramOutput` objects make results analysis easy!
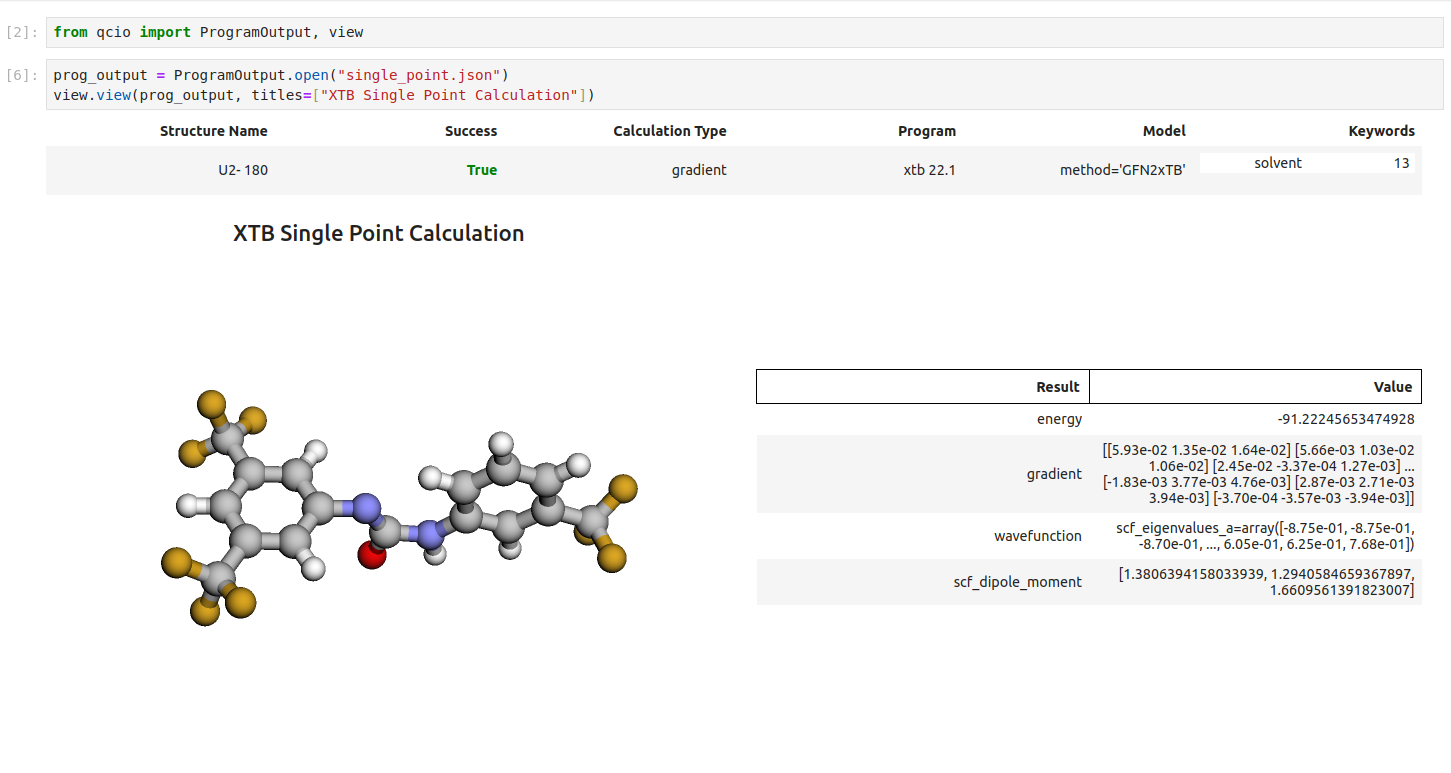
Single point calculations display their results in a table.
If you want to use the HTML generated by the viewer to build your own dashboards use the functions inside of `qcio.view.py` that begin with the word `generate_` to create HTML you can insert into any dashboard.
## Support
If you have any issues with `qcop` or would like to request a feature, please open an [issue](https://github.com/coltonbh/qcop/issues).
Raw data
{
"_id": null,
"home_page": null,
"name": "qcop",
"maintainer": null,
"docs_url": null,
"requires_python": "<4.0,>=3.9",
"maintainer_email": null,
"keywords": null,
"author": "Colton Hicks",
"author_email": "github@coltonhicks.com",
"download_url": "https://files.pythonhosted.org/packages/0d/25/2d70bce86b5f3d588850828e2c6736ea038e20e9e1bd1753e54ac3509c7a/qcop-0.9.8.tar.gz",
"platform": null,
"description": "# Quantum Chemistry Operate\n\nA package for operating Quantum Chemistry programs using [qcio](https://github.com/coltonbh/qcio) standardized data structures. Compatible with `TeraChem`, `psi4`, `QChem`, `NWChem`, `ORCA`, `Molpro`, `geomeTRIC` and many more.\n\n[](https://pypi.python.org/pypi/qcop)\n[](https://pypi.python.org/pypi/qcop)\n[](https://pypi.python.org/pypi/qcop)\n[](https://github.com/coltonbh/qcop/actions)\n[](https://github.com/coltonbh/qcop/actions)\n[](https://github.com/charliermarsh/ruff)\n\n`qcop` works in harmony with a suite of other quantum chemistry tools for fast, structured, and interoperable quantum chemistry.\n\n## The QC Suite of Programs\n\n- [qcio](https://github.com/coltonbh/qcio) - Elegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations.\n- [qcparse](https://github.com/coltonbh/qcparse) - A library for efficient parsing of quantum chemistry data into structured `qcio` objects.\n- [qcop](https://github.com/coltonbh/qcop) - A package for operating quantum chemistry programs using `qcio` standardized data structures. Compatible with `TeraChem`, `psi4`, `QChem`, `NWChem`, `ORCA`, `Molpro`, `geomeTRIC`, and many more, featuring seamless Jupyter Notebook visualizations.\n- [BigChem](https://github.com/mtzgroup/bigchem) - A distributed application for running quantum chemistry calculations at scale across clusters of computers or the cloud. Bring multi-node scaling to your favorite quantum chemistry program, featuring seamless Jupyter Notebook visualizations.\n- `ChemCloud` - A [web application](https://github.com/mtzgroup/chemcloud-server) and associated [Python client](https://github.com/mtzgroup/chemcloud-client) for exposing a BigChem cluster securely over the internet, featuring seamless Jupyter Notebook visualizations.\n\n## Installation\n\n```sh\npip install qcop\n```\n\n## Quickstart\n\n`qcop` uses the `qcio` data structures to drive quantum chemistry programs in a standardized way. This allows for a simple and consistent interface to a wide variety of quantum chemistry programs. See the [qcio](https://github.com/coltonbh/qcio) library for documentation on the input and output data structures.\n\nThe `compute` function is the main entry point for the library and is used to run a calculation.\n\n```python\nfrom qcio import Structure, ProgramInput\nfrom qcop import compute\nfrom qcop.exceptions import ExternalProgramError\n# Create the Structure\nh2o = Structure.open(\"h2o.xyz\")\n\n# Define the program input\nprog_input = ProgramInput(\n structure=h2o,\n calctype=\"energy\",\n model={\"method\": \"hf\", \"basis\": \"sto-3g\"},\n keywords={\"purify\": \"no\", \"restricted\": False},\n)\n\n# Run the calculation; will return ProgramOutput or raise an exception\ntry:\n po = compute(\"terachem\", prog_input, collect_files=True)\nexcept ExternalProgramError as e:\n # External QQ program failed in some way\n po = e.program_output\n po.input_data # Input data used by the QC program\n po.success # Will be False\n po.results # Any half-computed results before the failure\n po.traceback # Stack trace from the calculation\n po.ptraceback # Shortcut to print out the traceback in human readable format\n po.stdout # Stdout log from the calculation\n raise e\nelse:\n # Calculation succeeded\n po.input_data # Input data used by the QC program\n po.success # Will be True\n po.results # All structured results from the calculation\n po.stdout # Stdout log from the calculation\n po.pstdout # Shortcut to print out the stdout in human readable format\n po.files # Any files returned by the calculation\n po.provenance # Provenance information about the calculation\n po.extras # Any extra information not in the schema\n\n```\n\nOne may also call `compute(..., raise_exc=False)` to return a `ProgramOutput` object rather than raising an exception when a calculation fails. This may allow easier handling of failures in some cases.\n\n```python\nfrom qcio import Structure, ProgramInput\nfrom qcop import compute\nfrom qcop.exceptions import ExternalProgramError\n# Create the Structure\nh2o = Structure.open(\"h2o.xyz\")\n\n# Define the program input\nprog_input = ProgramInput(\n structure=h2o,\n calctype=\"energy\",\n model={\"method\": \"hf\", \"basis\": \"sto-3g\"},\n keywords={\"purify\": \"no\", \"restricted\": False},\n)\n\n# Run the calculation; will return a ProgramOutput objects\npo = compute(\"terachem\", prog_input, collect_files=True, raise_exc=False)\nif not po.success:\n # External QQ program failed in some way\n po.input_data # Input data used by the QC program\n po.success # Will be False\n po.results # Any half-computed results before the failure\n po.traceback # Stack trace from the calculation\n po.ptraceback # Shortcut to print out the traceback in human readable format\n po.stdout # Stdout log from the calculation\n\nelse:\n # Calculation succeeded\n po.input_data # Input data used by the QC program\n po.success # Will be True\n po.results # All structured results from the calculation\n po.stdout # Stdout log from the calculation\n po.pstdout # Shortcut to print out the stdout in human readable format\n po.files # Any files returned by the calculation\n po.provenance # Provenance information about the calculation\n po.extras # Any extra information not in the schema\n\n```\n\nAlternatively, the `compute_args` function can be used to run a calculation with the input data structures passed in as arguments rather than as a single `ProgramInput` object.\n\n```python\nfrom qcio import Structure\nfrom qcop import compute_args\n# Create the Structure\nh2o = Structure.open(\"h2o.xyz\")\n\n# Run the calculation\noutput = compute_args(\n \"terachem\",\n h2o,\n calctype=\"energy\",\n model={\"method\": \"hf\", \"basis\": \"sto-3g\"},\n keywords={\"purify\": \"no\", \"restricted\": False},\n files={...},\n collect_files=True\n)\n```\n\nThe behavior of `compute()` and `compute_args()` can be tuned by passing in keyword arguments like `collect_files` shown above. Keywords can modify which scratch directory location to use, whether to delete or keep the scratch files after a calculation completes, what files to collect from a calculation, whether to print the program stdout in real time as the program executes, and whether to propagate a wavefunction through a series of calculations. Keywords also include hooks for passing in update functions that can be called as a program executes in real time. See the [compute method docstring](https://github.com/coltonbh/qcop/blob/83868df51d241ffae3497981dfc3c72235319c6e/qcop/adapters/base.py#L57-L123) for more details.\n\nSee the [/examples](https://github.com/coltonbh/qcop/tree/master/examples) directory for more examples.\n\n## \u2728 Visualization \u2728\n\nVisualize all your results with a single line of code!\n\nFirst install the visualization module:\n\n```sh\npip install qcio[view]\n```\n\nor if your shell requires `''` around arguments with brackets:\n\n```sh\npip install 'qcio[view]'\n```\n\nThen in a Jupyter notebook import the `qcio` view module and call `view.view(...)` passing it one or any number of `qcio` objects you want to visualizing including `Structure` objects or any `ProgramOutput` object. You may also pass an array of `titles` and/or `subtitles` to add additional information to the molecular structure display. If no titles are passed `qcio` with look for `Structure` identifiers such as a name or SMILES to label the `Structure`.\n\n\n\nSeamless visualizations for `ProgramOutput` objects make results analysis easy!\n\n\n\nSingle point calculations display their results in a table.\n\n\n\nIf you want to use the HTML generated by the viewer to build your own dashboards use the functions inside of `qcio.view.py` that begin with the word `generate_` to create HTML you can insert into any dashboard.\n\n## Support\n\nIf you have any issues with `qcop` or would like to request a feature, please open an [issue](https://github.com/coltonbh/qcop/issues).\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A package for operating Quantum Chemistry programs using qcio standardized data structures. Compatible with TeraChem, psi4, QChem, NWChem, ORCA, Molpro, geomeTRIC and many more.",
"version": "0.9.8",
"project_urls": {
"Homepage": "https://github.com/coltonbh/qcop",
"Repository": "https://github.com/coltonbh/qcop"
},
"split_keywords": [],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "fe9edb3f580d7596075485b9cc1f78bea8ebc8bcc78f4919fe758b4ee143e698",
"md5": "2e71f4d9d75eefe0cb453a8a0fa7b3ad",
"sha256": "e2791c216496103b97bce589a8e9db392830101e08bcb424a1407c7ed3b8b1ed"
},
"downloads": -1,
"filename": "qcop-0.9.8-py3-none-any.whl",
"has_sig": false,
"md5_digest": "2e71f4d9d75eefe0cb453a8a0fa7b3ad",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": "<4.0,>=3.9",
"size": 29708,
"upload_time": "2025-03-01T00:58:37",
"upload_time_iso_8601": "2025-03-01T00:58:37.975193Z",
"url": "https://files.pythonhosted.org/packages/fe/9e/db3f580d7596075485b9cc1f78bea8ebc8bcc78f4919fe758b4ee143e698/qcop-0.9.8-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "0d252d70bce86b5f3d588850828e2c6736ea038e20e9e1bd1753e54ac3509c7a",
"md5": "700c6ee24d9cdfcfc3fb60b5e9f599fd",
"sha256": "4b323acc0d3c7f51dbef6e963c886988e1a5274abb5d4ae58b7093adf4775738"
},
"downloads": -1,
"filename": "qcop-0.9.8.tar.gz",
"has_sig": false,
"md5_digest": "700c6ee24d9cdfcfc3fb60b5e9f599fd",
"packagetype": "sdist",
"python_version": "source",
"requires_python": "<4.0,>=3.9",
"size": 24365,
"upload_time": "2025-03-01T00:58:40",
"upload_time_iso_8601": "2025-03-01T00:58:40.036523Z",
"url": "https://files.pythonhosted.org/packages/0d/25/2d70bce86b5f3d588850828e2c6736ea038e20e9e1bd1753e54ac3509c7a/qcop-0.9.8.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-03-01 00:58:40",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "coltonbh",
"github_project": "qcop",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "qcop"
}
