# Quasinet


<p align="center">
<img src="http://zed.uchicago.edu/logo/logozed1.png">
</p>
## Description
Infer non-local structural dependencies in genomic sequences. Genomic sequences are esentially compressed encodings of phenotypic information. This package provides a novel set of tools to extract long-range structural dependencies in genotypic data that define the phenotypic outcomes. The key capabilities implemented here are as follows:
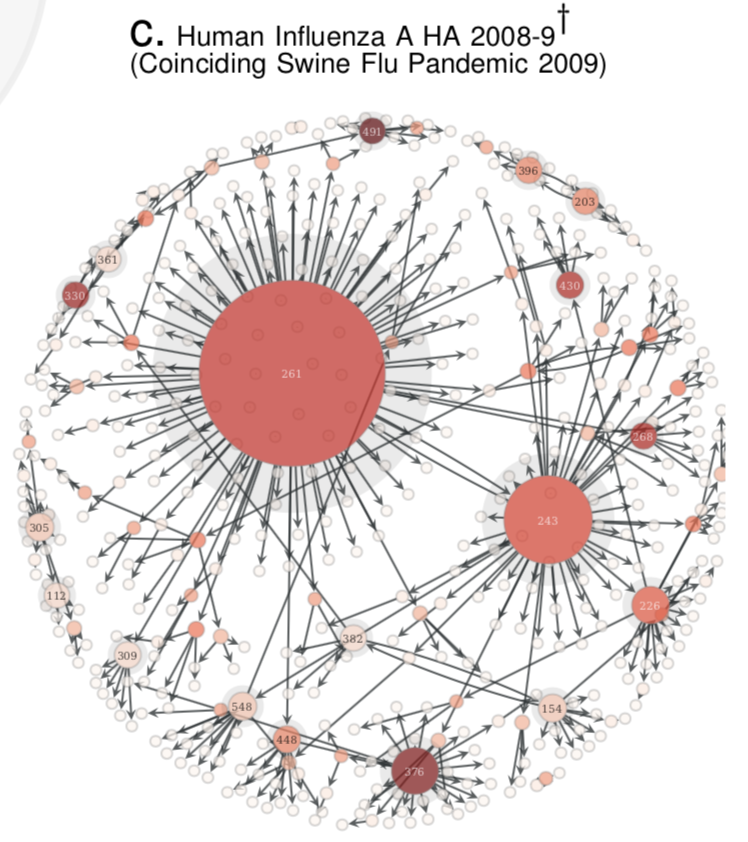
1. Compute the Quasinet (Q-net) given a database of nucleic acid sequences. The Q-net is a family of conditional inference trees that capture the predictability of each nucleotide position given the rest of the genome. The constructed Q-net for COVID-19 and Influenza A H1N1 HA 2008-9 is shown below.
COVID-19 | INFLUENZA
:-------------------------:|:-------------------------:
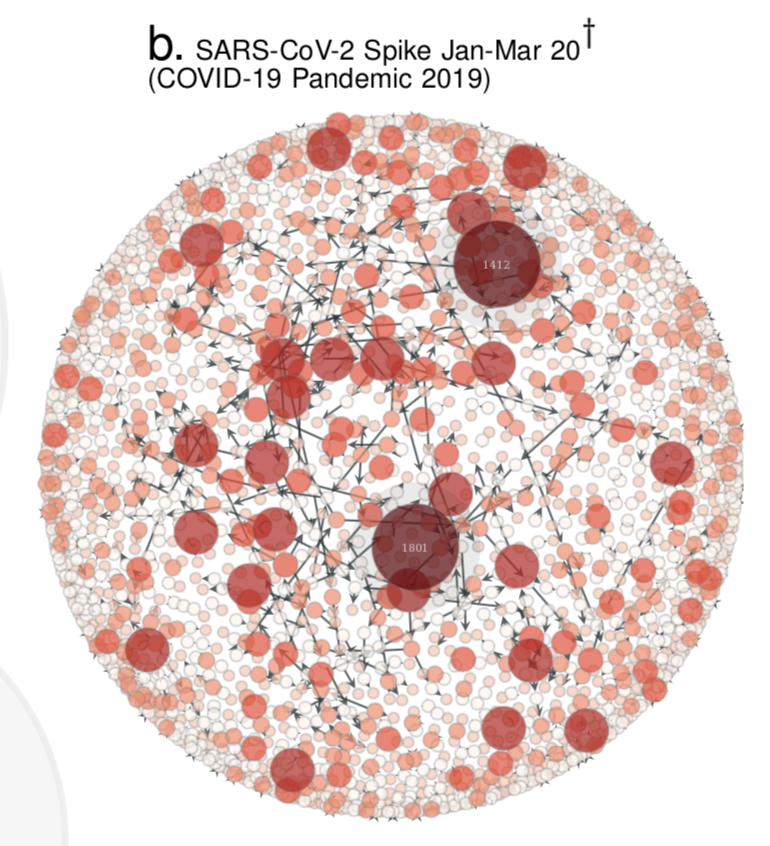 | 
2. Compute a structure-aware evolution-adaptive notion of distance between genomes, which is demonstrably more biologically relevant compared to the standard edit distance.
3. Draw samples in-silico that have a high probability of being biologically correct. For example, given a database of Influenza sequences, we can generate a new genomic sequence that has a high probability of being a valid influenza sequence.
<!-- { width=25% } -->
<p align="center">
<img src="https://raw.githubusercontent.com/zeroknowledgediscovery/quasinet/master/images/sampling.png" width="50%" height="50%">
</p>
## Installation
To install with pip:
```
pip install quasinet
```
To fix error with Mac or Windows:
```
from quasinet.osfix import osfix
# for windows
osfix('win')
# for max x86_64 (macbook pro)
osfix('macx86')
# mac arm (macbook air)
osfix('macarm')
```
NOTE: If trying to reproduce the paper below, please use `pip install quasinet==0.0.58`
### Dependencies
* scikit-learn
* scipy
* numpy
* numba
* pandas
* joblib
* biopython
## Usage
```
from quasinet import qnet
# initialize qnet
myqnet = qnet.Qnet()
# train the qnet
myqnet.fit(X)
# compute qdistance
qdist = qnet.qdistance(seq1, seq2, myqnet, myqnet)
```
### Examples
Examples are located [here](https://github.com/zeroknowledgediscovery/quasinet/tree/master/examples).
## Documentation
For more documentation, see [here](https://zeroknowledgediscovery.github.io/quasinet/build/html/index.html).
## Papers
For reference, please check out our paper:
[Preparing For the Next Pandemic: Learning Wild Mutational Patterns At Scale For Analyzing Sequence Divergence In Novel Pathogens](https://www.medrxiv.org/content/10.1101/2020.07.17.20156364v3)
## Authors
You can reach the ZED lab at: zed.uchicago.edu
Raw data
{
"_id": null,
"home_page": "https://github.com/zeroknowledgediscovery/",
"name": "quasinet",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.3",
"maintainer_email": null,
"keywords": "decision trees, machine learning, computational biology",
"author": "zed.uchicago.edu",
"author_email": "ishanu@uchicago.edu",
"download_url": "https://files.pythonhosted.org/packages/a3/0c/c266bf73bb43c6b4b3d0bf037fa6d12a27900f65290f2d41a2f6798e4ce4/quasinet-0.1.71.tar.gz",
"platform": null,
"description": "# Quasinet\n\n\n\n\n\n\n<p align=\"center\">\n <img src=\"http://zed.uchicago.edu/logo/logozed1.png\">\n</p>\n\n\n## Description\n\nInfer non-local structural dependencies in genomic sequences. Genomic sequences are esentially compressed encodings of phenotypic information. This package provides a novel set of tools to extract long-range structural dependencies in genotypic data that define the phenotypic outcomes. The key capabilities implemented here are as follows: \n\n1. Compute the Quasinet (Q-net) given a database of nucleic acid sequences. The Q-net is a family of conditional inference trees that capture the predictability of each nucleotide position given the rest of the genome. The constructed Q-net for COVID-19 and Influenza A H1N1 HA 2008-9 is shown below.\n\nCOVID-19 | INFLUENZA\n:-------------------------:|:-------------------------:\n | \n\n\n\n2. Compute a structure-aware evolution-adaptive notion of distance between genomes, which is demonstrably more biologically relevant compared to the standard edit distance. \n\n3. Draw samples in-silico that have a high probability of being biologically correct. For example, given a database of Influenza sequences, we can generate a new genomic sequence that has a high probability of being a valid influenza sequence.\n\n<!-- { width=25% } -->\n\n<p align=\"center\">\n <img src=\"https://raw.githubusercontent.com/zeroknowledgediscovery/quasinet/master/images/sampling.png\" width=\"50%\" height=\"50%\">\n</p>\n\n## Installation\n\nTo install with pip:\n\n```\npip install quasinet\n```\n\nTo fix error with Mac or Windows:\n\n```\nfrom quasinet.osfix import osfix\n# for windows\nosfix('win')\n# for max x86_64 (macbook pro)\nosfix('macx86')\n# mac arm (macbook air)\nosfix('macarm')\n\n```\n\n\n\nNOTE: If trying to reproduce the paper below, please use `pip install quasinet==0.0.58`\n\n### Dependencies\n\n* scikit-learn \n* scipy \n* numpy \n* numba \n* pandas \n* joblib \n* biopython\n\n## Usage\n\n```\nfrom quasinet import qnet\n\n# initialize qnet\nmyqnet = qnet.Qnet()\n\n# train the qnet\nmyqnet.fit(X)\n\n# compute qdistance\nqdist = qnet.qdistance(seq1, seq2, myqnet, myqnet) \n```\n \n### Examples\n\nExamples are located [here](https://github.com/zeroknowledgediscovery/quasinet/tree/master/examples).\n\n## Documentation\n\nFor more documentation, see [here](https://zeroknowledgediscovery.github.io/quasinet/build/html/index.html).\n\n## Papers\n\nFor reference, please check out our paper: \n\n[Preparing For the Next Pandemic: Learning Wild Mutational Patterns At Scale For Analyzing Sequence Divergence In Novel Pathogens](https://www.medrxiv.org/content/10.1101/2020.07.17.20156364v3)\n\n## Authors\n\nYou can reach the ZED lab at: zed.uchicago.edu\n",
"bugtrack_url": null,
"license": "LICENSE.txt",
"summary": "Utitilies for constructing and manipulating models for non-local structural dependencies in genomic sequences",
"version": "0.1.71",
"project_urls": {
"Download": "https://github.com/zeroknowledgediscovery/quasinet/archive/0.1.71.tar.gz",
"Homepage": "https://github.com/zeroknowledgediscovery/"
},
"split_keywords": [
"decision trees",
" machine learning",
" computational biology"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "1a269df55b6362bbc2e433d8180519b610586cc694d544a484ee2bc65b45c72d",
"md5": "beebc54a97cf9a0c7a7d25952140d7bd",
"sha256": "7a816fdf261deb7552372c30dfe77f10fc6c3e6d0eac9a239156ec75d7d374b4"
},
"downloads": -1,
"filename": "quasinet-0.1.71-py3-none-any.whl",
"has_sig": false,
"md5_digest": "beebc54a97cf9a0c7a7d25952140d7bd",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.3",
"size": 15234760,
"upload_time": "2025-01-20T21:32:02",
"upload_time_iso_8601": "2025-01-20T21:32:02.958319Z",
"url": "https://files.pythonhosted.org/packages/1a/26/9df55b6362bbc2e433d8180519b610586cc694d544a484ee2bc65b45c72d/quasinet-0.1.71-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "a30cc266bf73bb43c6b4b3d0bf037fa6d12a27900f65290f2d41a2f6798e4ce4",
"md5": "9cd542a619a60f40af4e09ab9dee8c05",
"sha256": "28c2eaa7fd02176231015b3e028130da6f32425ecee01bd63d9242def5139ff4"
},
"downloads": -1,
"filename": "quasinet-0.1.71.tar.gz",
"has_sig": false,
"md5_digest": "9cd542a619a60f40af4e09ab9dee8c05",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.3",
"size": 14483567,
"upload_time": "2025-01-20T21:32:06",
"upload_time_iso_8601": "2025-01-20T21:32:06.763674Z",
"url": "https://files.pythonhosted.org/packages/a3/0c/c266bf73bb43c6b4b3d0bf037fa6d12a27900f65290f2d41a2f6798e4ce4/quasinet-0.1.71.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-01-20 21:32:06",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "zeroknowledgediscovery",
"github_project": "quasinet",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "quasinet"
}
