| Name | ramachandraw JSON |
| Version |
1.0.1
 JSON
JSON |
| download |
| home_page | |
| Summary | Ramachandran plotting tool |
| upload_time | 2024-02-29 22:10:16 |
| maintainer | |
| docs_url | None |
| author | Alexandre Cirilo |
| requires_python | >=3.9,<4.0 |
| license | MIT |
| keywords |
|
| VCS |
|
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
# Ramachandran plotting tool
[](https://github.com/alxdrcirilo/ramachandraw/actions/workflows/coveralls.yaml)
[](https://coveralls.io/github/alxdrcirilo/ramachandraw?branch=master)
[](https://www.python.org/)
[](https://python-poetry.org/)
[](https://github.com/astral-sh/ruff)
[](https://opensource.org/licenses/MIT)


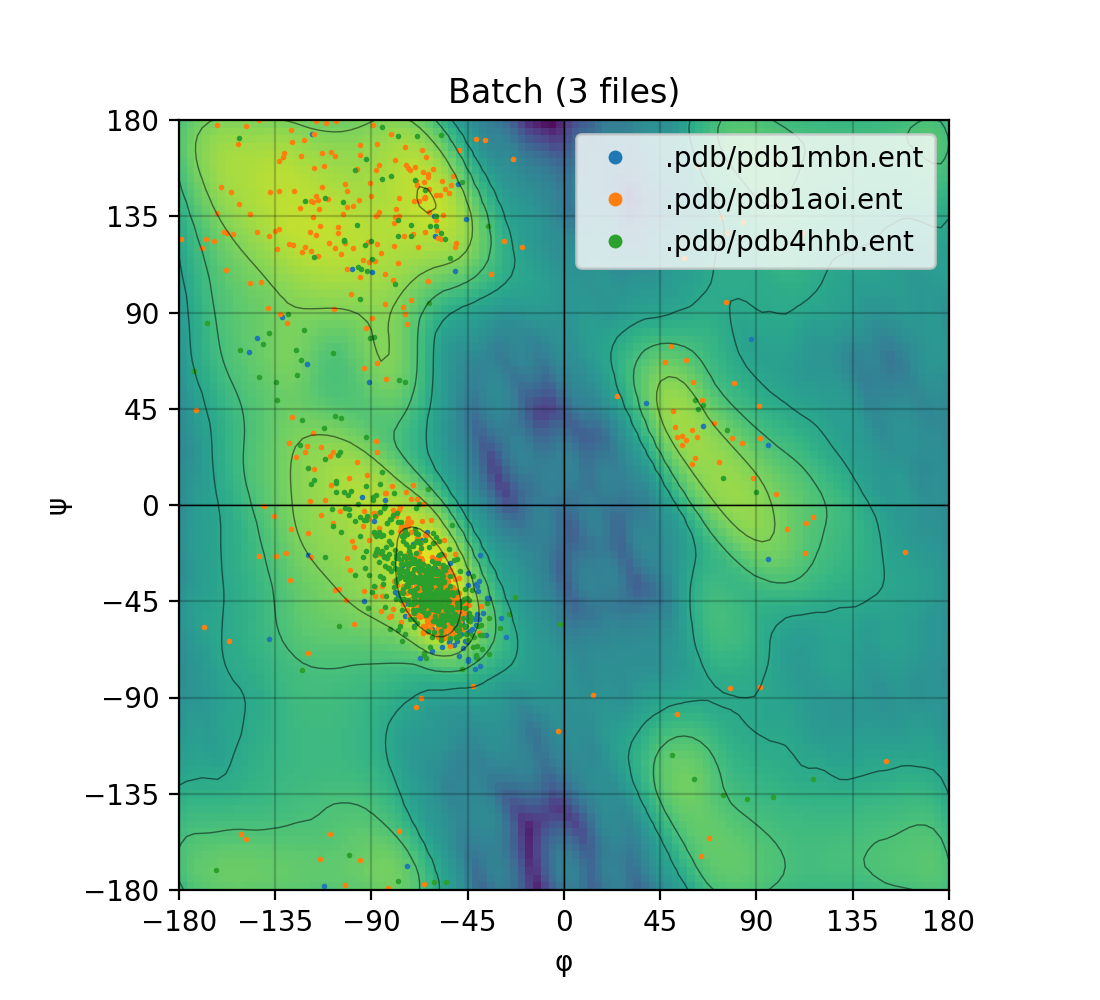
Draws a [Ramachandran plot](https://en.wikipedia.org/wiki/Ramachandran_plot) based on the input PDB file (e.g. `1mbn.pdb`). Makes use of a Gaussian KDE (kernel density
estimation) to plot the density of favoured torsion angles (φ and ψ).
Single mode | Batch mode
:-------------------------:|:-------------------------:
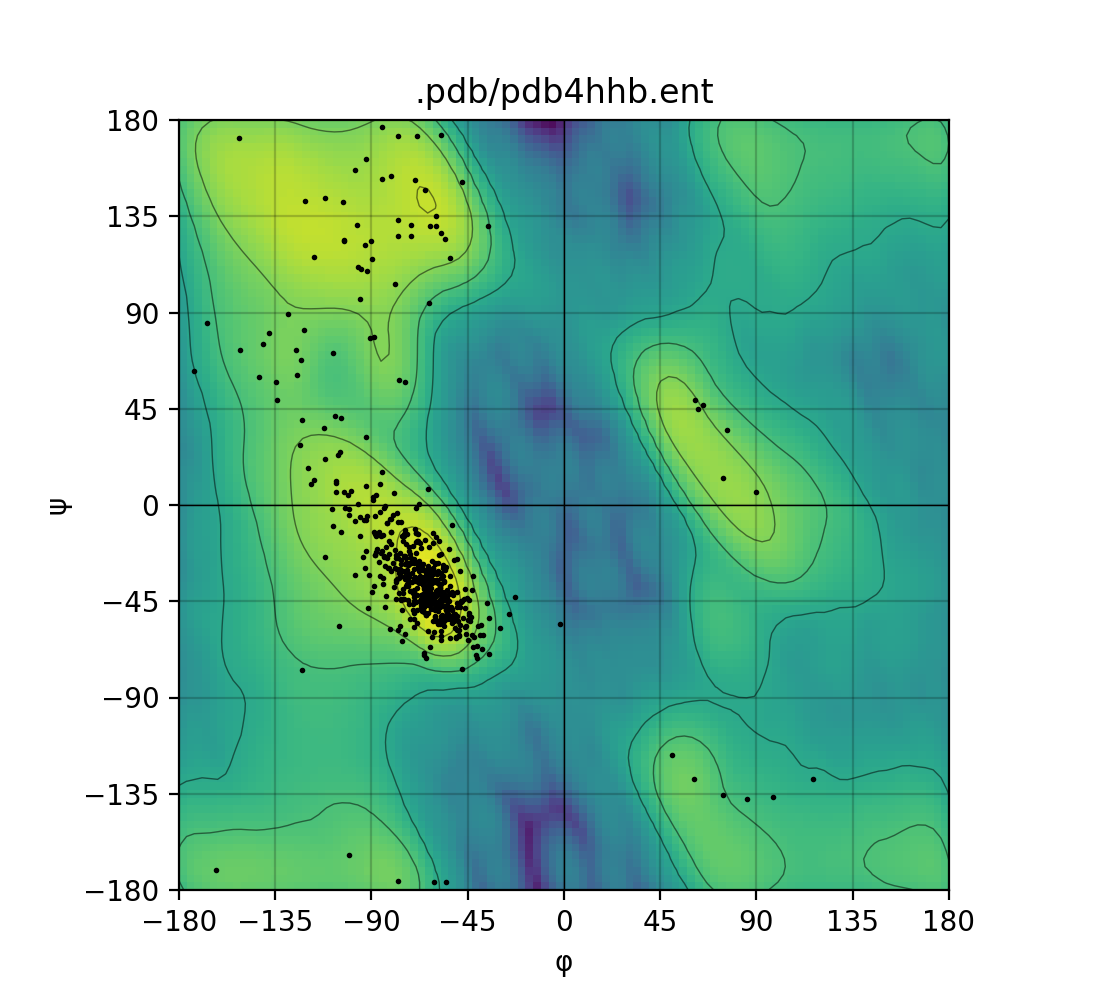 | 
## Installation
RamachanDraw is hosted on [PyPi](https://pypi.org/project/RamachanDraw/).
```bash
pip install ramachandraw
```
## Usage
RamachanDraw includes useful functions to effortlessly draw a Ramachandran plot.
### Fetch the PDB file
To draw a Ramachandran plot, we need a PDB file. RamachanDraw conveniently includes a function to automatically fetch and locally store the PDB file for the given PDB id.
#### Example
```python
from ramachandraw.utils import fetch_pdb
fetch_pdb(pdb_id, verbose)
```
- `pdb_id (str|list|tuple)`: PDB id(s) corresponding to the PDB file(s) to be downloaded
- `verbose (bool)` (*optional*): set the verbosity, defaults to `False`
- **Returns**: path(s) to PDB file(s)
### Extract φ and ψ torsion angles
RamachanDraw extracts the φ and ψ torsion angles from the PDB file by taking advantage of the [Biopython](https://biopython.org/) module. Additionally, aminoacid residues that were not drawn on the plot can be extract using the `return_ignored` argument.
#### Example
```python
from ramachandraw.parser import get_phi_psi
phi_psi(pdb_filepath, prune, hide_warnings)
```
- `pdb_id (str|list|tuple)`: PDB filepath(s)
- `prune (bool)` (*optional*): prunes aminoacids with missing torsion angle(s), defaults to `True`
- `hide_warnings (bool)` (*optional*): sets the verbosity of the PDB structure parser, defaults to `True`
- **Returns**: Dictionary with keys as aminoacid residues and values as (φ, ψ) angles.
### Ramachandran plot
Makes use of the [matplotlib](https://matplotlib.org/) module to draw a highly customizable Ramachandran plot.
#### Example
```python
from ramachandraw.utils import plot
plot(pdb_filepath, cmap="viridis", alpha=0.75, dpi=100, save=True, show=False, filename="plot.png")
```
- `pdb_file (str|list|tuple)`: PDB id(s) corresponding to the PDB entry to be downloaded
- `cmap (str)` (*optional*): colormap to be used (from `matplotlib`) for the density of the favoured ("allowed") regions; default
is *viridis*.
- `alpha (float)` (*optional*): sets the opacity of the colormap (value between 0-1); default is 0.75.
- `dpi (int)` (*optional*): resolution (in *dots per inch*); default is `100`.
- `save (bool)` (*optional*):
- `True`: saves the plot locally; default is True.
- `show (bool)` (*optional*):
- `True`: shows the plot using the Qt5Agg backend; default is False.
- `filename (str)` (*optional*): filename to be used in case the plot is saved (i.e. `save=True`); default is `plot.png`.
- **Returns**: Ramachandran plot (`matplotlib.axes.Axes` object) that can be further customized if needed
## Example
Herein you will find an example from the PDB id corresponding to the myoglobin entry: [1MBN](https://www.ebi.ac.uk/pdbe/entry/pdb/1mbn/index) - in the Protein Data Bank.
```python
from ramachandraw.parser import get_phi_psi
from ramachandraw.utils import fetch_pdb, plot
# PDB id
pdb_id = "1mbn"
# Draw the Ramachandran plot
plot(fetch_pdb(pdb_id))
# Generate a dictionary to store the (phi, psi) torsion angles
torsion_angles = get_phi_psi(fetch_pdb(pdb_id))
```
## Contributing
Feedback and constructive criticism is welcome. If necessary, open an issue in the [issues](https://github.com/alxdrcirilo/ramachandraw/issues) tab.
## License
[MIT](https://choosealicense.com/licenses/mit/)
Raw data
{
"_id": null,
"home_page": "",
"name": "ramachandraw",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.9,<4.0",
"maintainer_email": "",
"keywords": "",
"author": "Alexandre Cirilo",
"author_email": "38657258+alxdrcirilo@users.noreply.github.com",
"download_url": "https://files.pythonhosted.org/packages/4f/69/217c70a33e785dcb0752f1b75f115708b95031cb1184a23a0aad45f5c9ec/ramachandraw-1.0.1.tar.gz",
"platform": null,
"description": "# Ramachandran plotting tool\n\n[](https://github.com/alxdrcirilo/ramachandraw/actions/workflows/coveralls.yaml)\n[](https://coveralls.io/github/alxdrcirilo/ramachandraw?branch=master)\n[](https://www.python.org/)\n[](https://python-poetry.org/)\n[](https://github.com/astral-sh/ruff)\n[](https://opensource.org/licenses/MIT)\n\n\n\nDraws a [Ramachandran plot](https://en.wikipedia.org/wiki/Ramachandran_plot) based on the input PDB file (e.g. `1mbn.pdb`). Makes use of a Gaussian KDE (kernel density\nestimation) to plot the density of favoured torsion angles (φ and ψ).\n\nSingle mode | Batch mode\n:-------------------------:|:-------------------------:\n | \n\n## Installation\n\nRamachanDraw is hosted on [PyPi](https://pypi.org/project/RamachanDraw/).\n\n```bash\npip install ramachandraw\n```\n\n## Usage\n\nRamachanDraw includes useful functions to effortlessly draw a Ramachandran plot.\n\n### Fetch the PDB file\n\nTo draw a Ramachandran plot, we need a PDB file. RamachanDraw conveniently includes a function to automatically fetch and locally store the PDB file for the given PDB id.\n\n#### Example\n\n```python\nfrom ramachandraw.utils import fetch_pdb\n\nfetch_pdb(pdb_id, verbose)\n```\n\n- `pdb_id (str|list|tuple)`: PDB id(s) corresponding to the PDB file(s) to be downloaded\n- `verbose (bool)` (*optional*): set the verbosity, defaults to `False`\n- **Returns**: path(s) to PDB file(s)\n\n### Extract φ and ψ torsion angles\n\nRamachanDraw extracts the φ and ψ torsion angles from the PDB file by taking advantage of the [Biopython](https://biopython.org/) module. Additionally, aminoacid residues that were not drawn on the plot can be extract using the `return_ignored` argument.\n\n#### Example\n\n```python\nfrom ramachandraw.parser import get_phi_psi\n\nphi_psi(pdb_filepath, prune, hide_warnings)\n```\n\n- `pdb_id (str|list|tuple)`: PDB filepath(s)\n- `prune (bool)` (*optional*): prunes aminoacids with missing torsion angle(s), defaults to `True`\n- `hide_warnings (bool)` (*optional*): sets the verbosity of the PDB structure parser, defaults to `True`\n- **Returns**: Dictionary with keys as aminoacid residues and values as (φ, ψ) angles.\n\n### Ramachandran plot\n\nMakes use of the [matplotlib](https://matplotlib.org/) module to draw a highly customizable Ramachandran plot.\n\n#### Example\n\n```python\nfrom ramachandraw.utils import plot\n\nplot(pdb_filepath, cmap=\"viridis\", alpha=0.75, dpi=100, save=True, show=False, filename=\"plot.png\")\n```\n\n- `pdb_file (str|list|tuple)`: PDB id(s) corresponding to the PDB entry to be downloaded\n- `cmap (str)` (*optional*): colormap to be used (from `matplotlib`) for the density of the favoured (\"allowed\") regions; default\n is *viridis*.\n- `alpha (float)` (*optional*): sets the opacity of the colormap (value between 0-1); default is 0.75.\n- `dpi (int)` (*optional*): resolution (in *dots per inch*); default is `100`.\n- `save (bool)` (*optional*):\n - `True`: saves the plot locally; default is True.\n- `show (bool)` (*optional*):\n - `True`: shows the plot using the Qt5Agg backend; default is False.\n- `filename (str)` (*optional*): filename to be used in case the plot is saved (i.e. `save=True`); default is `plot.png`.\n- **Returns**: Ramachandran plot (`matplotlib.axes.Axes` object) that can be further customized if needed\n\n## Example\n\nHerein you will find an example from the PDB id corresponding to the myoglobin entry: [1MBN](https://www.ebi.ac.uk/pdbe/entry/pdb/1mbn/index) - in the Protein Data Bank.\n\n```python\nfrom ramachandraw.parser import get_phi_psi\nfrom ramachandraw.utils import fetch_pdb, plot\n\n# PDB id\npdb_id = \"1mbn\"\n\n# Draw the Ramachandran plot\nplot(fetch_pdb(pdb_id))\n\n# Generate a dictionary to store the (phi, psi) torsion angles\ntorsion_angles = get_phi_psi(fetch_pdb(pdb_id))\n\n```\n\n## Contributing\n\nFeedback and constructive criticism is welcome. If necessary, open an issue in the [issues](https://github.com/alxdrcirilo/ramachandraw/issues) tab.\n\n## License\n\n[MIT](https://choosealicense.com/licenses/mit/)\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Ramachandran plotting tool",
"version": "1.0.1",
"project_urls": null,
"split_keywords": [],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "d1984de5516ce9ed3f296aa75f5c4919fd1df0aebbd93769ca34c2896cae6112",
"md5": "e606c05f7b68d1b194e4ec0275719dd2",
"sha256": "333b1baa22edee0e1faefdbcced5d4b05442d18c4a4da37cb67bfd6bfd75068a"
},
"downloads": -1,
"filename": "ramachandraw-1.0.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "e606c05f7b68d1b194e4ec0275719dd2",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.9,<4.0",
"size": 83808,
"upload_time": "2024-02-29T22:10:13",
"upload_time_iso_8601": "2024-02-29T22:10:13.216955Z",
"url": "https://files.pythonhosted.org/packages/d1/98/4de5516ce9ed3f296aa75f5c4919fd1df0aebbd93769ca34c2896cae6112/ramachandraw-1.0.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "4f69217c70a33e785dcb0752f1b75f115708b95031cb1184a23a0aad45f5c9ec",
"md5": "b1a1de64ff34ac48bb820dfcd8f18793",
"sha256": "1ce2cbdbe4457c2cdfab05c7ca14458d0b3b10916afd18b01642eec0f1e8f8de"
},
"downloads": -1,
"filename": "ramachandraw-1.0.1.tar.gz",
"has_sig": false,
"md5_digest": "b1a1de64ff34ac48bb820dfcd8f18793",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.9,<4.0",
"size": 85084,
"upload_time": "2024-02-29T22:10:16",
"upload_time_iso_8601": "2024-02-29T22:10:16.476601Z",
"url": "https://files.pythonhosted.org/packages/4f/69/217c70a33e785dcb0752f1b75f115708b95031cb1184a23a0aad45f5c9ec/ramachandraw-1.0.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-02-29 22:10:16",
"github": false,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"lcname": "ramachandraw"
}