# Rapid QC-MS
**Rapid QC-MS** is an all-in-one solution for automated quality control of liquid chromatography-mass spectrometry (LC-MS) instrument runs, both during and after data acquisition.
It offers a fast, straightforward approach to ensure collection of high-quality data, allowing for less time investigating raw data and more time conducting experiments.
Developed at the [Mass Spectrometry Platform of CZ Biohub San Francisco](https://www.czbiohub.org/mass-spec/), Rapid QC-MS provides a host of key features to streamline untargeted metabolomics research, such as:
- **Automated and user-defined quality control checks** during instrument runs
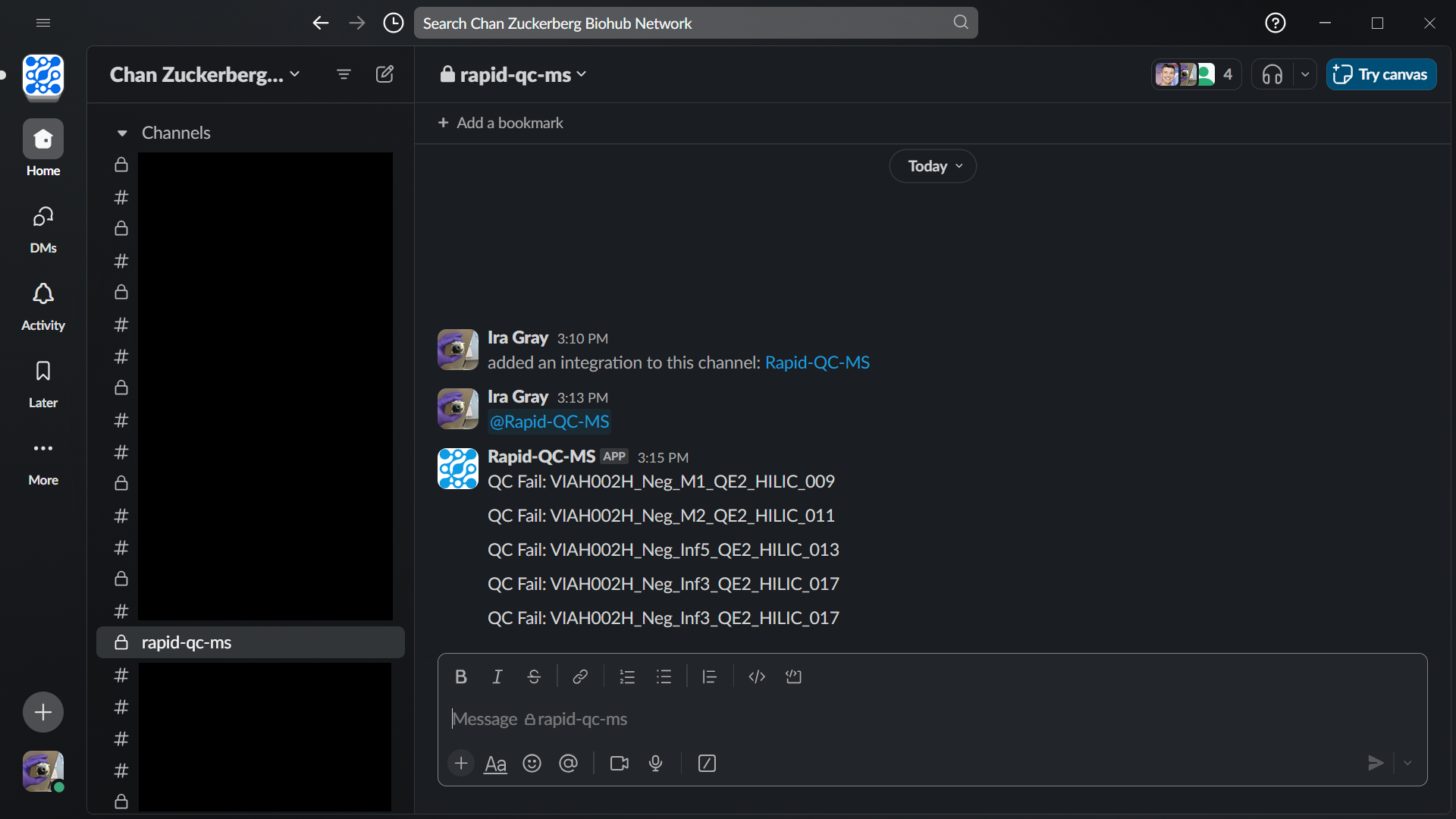
- **Realtime updates on QC fails** in the form of Slack or email notifications
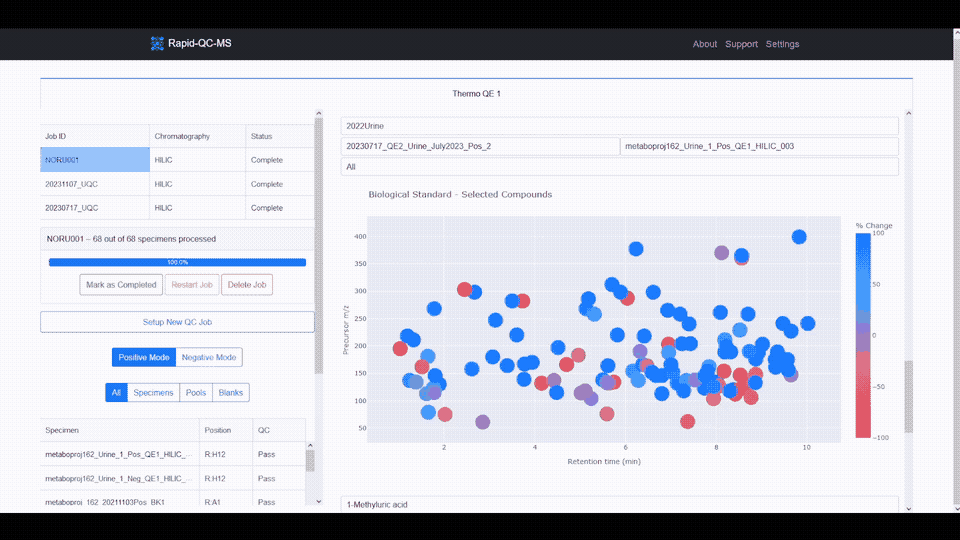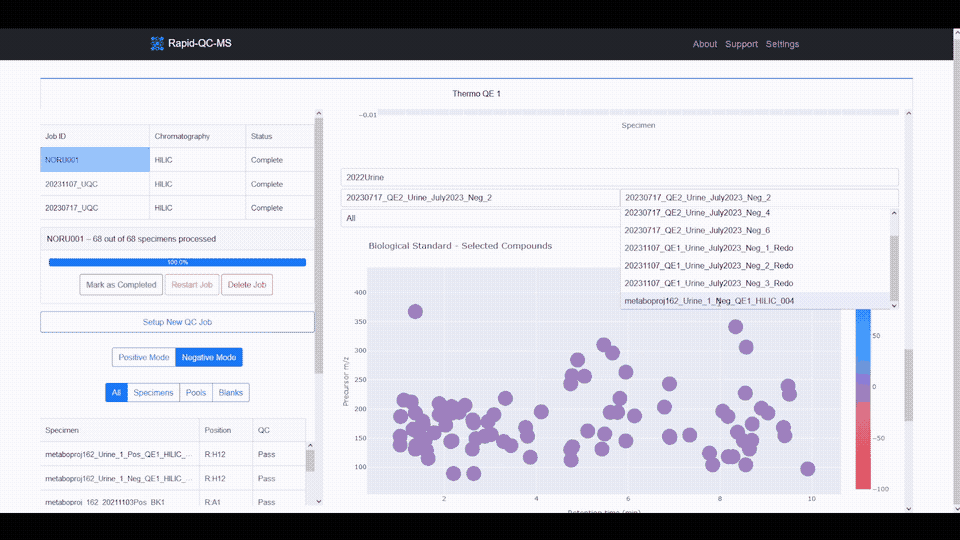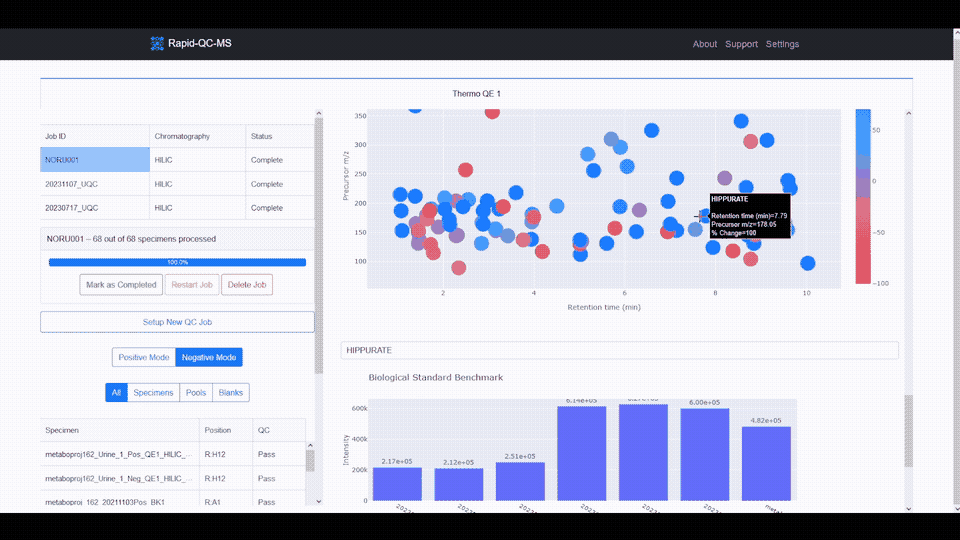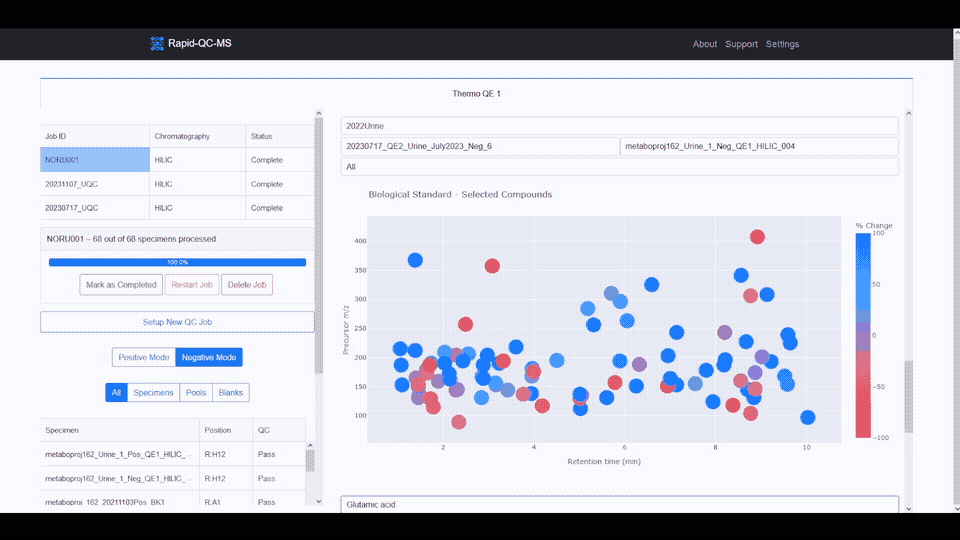
- **Interactive data visualization** of internal standard retention time, _m/z_, and intensity across samples
- **Google Drive cloud sync** and secure, Google-authenticated access to QC results from any device

# Requirements
**Rapid QC-MS was designed to run on Windows platforms** because of its dependency on [MSConvert](https://proteowizard.sourceforge.io/tools/msconvert.html) for vendor format data conversion and [MS-DIAL](http://prime.psc.riken.jp/compms/msdial/main.html) for data processing and identification. However, MacOS users can still use Rapid QC-MS to monitor / view their instrument run data.
In addition, Rapid QC-MS requires Python 3.8 to 3.11 and various Python packages, including:
- Pandas
- SQLAlchemy
- Plotly Dash
- Bootstrap
- Watchdog
- Google API
- Slack API
These are installed automatically during setup.
**Note:** Installation of Python and various Python packages on MS instrument computers comes at no risk. For extra security and peace of mind, you can opt to install Rapid QC-MS in a virtual environment. To learn more, please read the [installation guide](https://czbiohub-sf.github.io/Rapid-QC-MS/installation.html#2-install-ms-autoqc).
# Installation
Installing Rapid QC-MS is easy. Simply open your Terminal or Command Prompt and enter:
```python
py -m pip install rapid-qc-ms
```
If you have multiple versions of python on your system, you can specifiy the version you want to use this way
```py -3.# -m pip install rapid-qc-ms```
Python dependencies are installed automatically, but dependencies such as MSConvert and MS-DIAL will need to be installed manually.
You can also opt to download and install Rapid QC-MS manually, or in a virtual environment if you prefer. Check out the [installation guide](https://czbiohub-sf.github.io/Rapid-QC-MS/installation.html#2-install-ms-autoqc) for more details.
[Example data](http://dx.doi.org/10.21228/M8Z119) can be downloaded from metabolomics workbench with [supplementary example files](https://doi.org/10.5281/zenodo.10525183) (sequence .csv, library .msp, and additional .raw files not published through workbench) available on zenodo.
# Usage
To start Rapid QC-MS, simply enter:
```python
rapidqcms
```
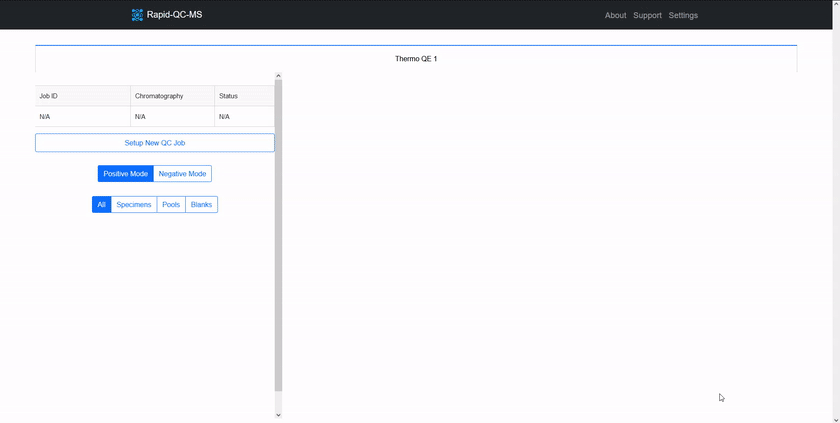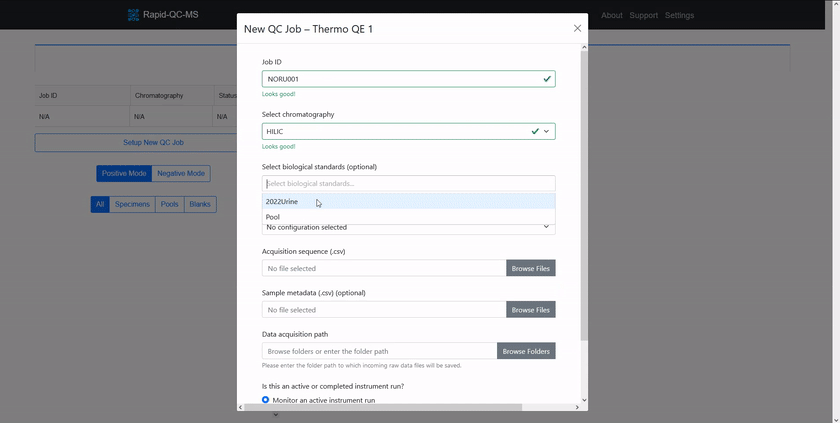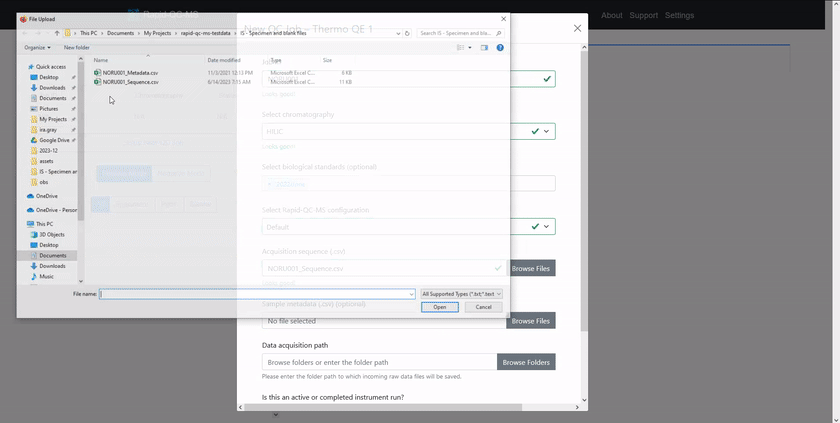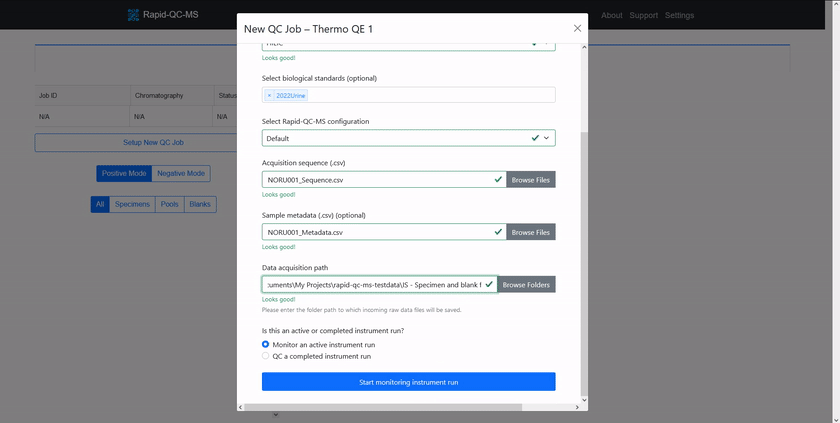
Check out the [quickstart guide](https://czbiohub-sf.github.io/Rapid-QC-MS/quickstart.html) to learn how easy setting up new QC jobs is.
# Supported instrument vendors
Rapid QC-MS was designed to be a universal, open-source solution for data quality control. Because MSConvert converts raw acquired data into open mzML format before routing it to the data processing pipeline, the package will work seamlessly with data of all vendor formats.
**However, Rapid QC-MS has only been tested extensively on Thermo Fisher mass spectrometers, Thermo acquisition sequences, and Thermo RAW files.** As such, it is expected that there may be bugs and issues with processing data of other vendor formats.
If you encounter a bug, please report it by [opening an issue on GitHub](https://github.com/czbiohub-sf/Rapid-QC-MS/issues).
We are open to collaboration! If you would like to help us develop support for Agilent, Bruker, Sciex, or Waters acquisition sequences and data files, please send an email to [brian.defelice@czbiohub.org](mailto:brian.defelice@czbiohub.org).
Raw data
{
"_id": null,
"home_page": null,
"name": "rapid-qc-ms",
"maintainer": null,
"docs_url": null,
"requires_python": "<3.12,>=3.8",
"maintainer_email": null,
"keywords": "mass-spectrometry, mass-spec, autoqc, mass-spec-quality-control",
"author": null,
"author_email": "Brian DeFelice <brian.defelice@czbiohub.org>",
"download_url": "https://files.pythonhosted.org/packages/a7/92/1cae19960134df7d1058bfdcc98906971ecba6b712933c8c4191725d58bd/rapid_qc_ms-1.1.0.tar.gz",
"platform": null,
"description": "# Rapid QC-MS\r\n**Rapid QC-MS** is an all-in-one solution for automated quality control of liquid chromatography-mass spectrometry (LC-MS) instrument runs, both during and after data acquisition.\r\n\r\n\r\n\r\nIt offers a fast, straightforward approach to ensure collection of high-quality data, allowing for less time investigating raw data and more time conducting experiments.\r\n\r\nDeveloped at the [Mass Spectrometry Platform of CZ Biohub San Francisco](https://www.czbiohub.org/mass-spec/), Rapid QC-MS provides a host of key features to streamline untargeted metabolomics research, such as:\r\n\r\n- **Automated and user-defined quality control checks** during instrument runs\r\n- **Realtime updates on QC fails** in the form of Slack or email notifications\r\n- **Interactive data visualization** of internal standard retention time, _m/z_, and intensity across samples\r\n- **Google Drive cloud sync** and secure, Google-authenticated access to QC results from any device\r\n\r\n\r\n\r\n\r\n\r\n# Requirements\r\n**Rapid QC-MS was designed to run on Windows platforms** because of its dependency on [MSConvert](https://proteowizard.sourceforge.io/tools/msconvert.html) for vendor format data conversion and [MS-DIAL](http://prime.psc.riken.jp/compms/msdial/main.html) for data processing and identification. However, MacOS users can still use Rapid QC-MS to monitor / view their instrument run data.\r\n\r\nIn addition, Rapid QC-MS requires Python 3.8 to 3.11 and various Python packages, including:\r\n\r\n- Pandas\r\n- SQLAlchemy\r\n- Plotly Dash\r\n- Bootstrap\r\n- Watchdog\r\n- Google API\r\n- Slack API\r\n\r\nThese are installed automatically during setup.\r\n\r\n**Note:** Installation of Python and various Python packages on MS instrument computers comes at no risk. For extra security and peace of mind, you can opt to install Rapid QC-MS in a virtual environment. To learn more, please read the [installation guide](https://czbiohub-sf.github.io/Rapid-QC-MS/installation.html#2-install-ms-autoqc).\r\n\r\n# Installation\r\nInstalling Rapid QC-MS is easy. Simply open your Terminal or Command Prompt and enter:\r\n```python\r\npy -m pip install rapid-qc-ms\r\n```\r\n\r\nIf you have multiple versions of python on your system, you can specifiy the version you want to use this way\r\n```py -3.# -m pip install rapid-qc-ms```\r\n\r\nPython dependencies are installed automatically, but dependencies such as MSConvert and MS-DIAL will need to be installed manually.\r\n\r\nYou can also opt to download and install Rapid QC-MS manually, or in a virtual environment if you prefer. Check out the [installation guide](https://czbiohub-sf.github.io/Rapid-QC-MS/installation.html#2-install-ms-autoqc) for more details.\r\n\r\n[Example data](http://dx.doi.org/10.21228/M8Z119) can be downloaded from metabolomics workbench with [supplementary example files](https://doi.org/10.5281/zenodo.10525183) (sequence .csv, library .msp, and additional .raw files not published through workbench) available on zenodo.\r\n\r\n# Usage\r\nTo start Rapid QC-MS, simply enter:\r\n```python\r\nrapidqcms\r\n```\r\n\r\nCheck out the [quickstart guide](https://czbiohub-sf.github.io/Rapid-QC-MS/quickstart.html) to learn how easy setting up new QC jobs is.\r\n\r\n\r\n\r\n# Supported instrument vendors\r\nRapid QC-MS was designed to be a universal, open-source solution for data quality control. Because MSConvert converts raw acquired data into open mzML format before routing it to the data processing pipeline, the package will work seamlessly with data of all vendor formats.\r\n\r\n**However, Rapid QC-MS has only been tested extensively on Thermo Fisher mass spectrometers, Thermo acquisition sequences, and Thermo RAW files.** As such, it is expected that there may be bugs and issues with processing data of other vendor formats.\r\n\r\nIf you encounter a bug, please report it by [opening an issue on GitHub](https://github.com/czbiohub-sf/Rapid-QC-MS/issues).\r\n\r\nWe are open to collaboration! If you would like to help us develop support for Agilent, Bruker, Sciex, or Waters acquisition sequences and data files, please send an email to [brian.defelice@czbiohub.org](mailto:brian.defelice@czbiohub.org).\r\n",
"bugtrack_url": null,
"license": null,
"summary": "Realtime quality control for mass spectrometry data acquisition",
"version": "1.1.0",
"project_urls": {
"Homepage": "https://czbiohub-sf.github.io/Rapid-QC-MS"
},
"split_keywords": [
"mass-spectrometry",
" mass-spec",
" autoqc",
" mass-spec-quality-control"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "8ffe15750861c5328fbd42c9ec97f4c454ce4dd867ac52dba736d3234c2bff1e",
"md5": "f5ca379e9bc6002eb3aa1721cc134d65",
"sha256": "539bb4f5011303b5165d4c6577cf8ddbc129c0e6ccb82920a586de3636e23086"
},
"downloads": -1,
"filename": "rapid_qc_ms-1.1.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "f5ca379e9bc6002eb3aa1721cc134d65",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": "<3.12,>=3.8",
"size": 145268,
"upload_time": "2024-07-01T21:31:11",
"upload_time_iso_8601": "2024-07-01T21:31:11.928813Z",
"url": "https://files.pythonhosted.org/packages/8f/fe/15750861c5328fbd42c9ec97f4c454ce4dd867ac52dba736d3234c2bff1e/rapid_qc_ms-1.1.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "a7921cae19960134df7d1058bfdcc98906971ecba6b712933c8c4191725d58bd",
"md5": "f7e3fddd92f2f89d6cb1e9fb6b13b770",
"sha256": "8a662950b1c80b1b197f27b9ca724834a8df78df7b4aa40e8664740284d48c2d"
},
"downloads": -1,
"filename": "rapid_qc_ms-1.1.0.tar.gz",
"has_sig": false,
"md5_digest": "f7e3fddd92f2f89d6cb1e9fb6b13b770",
"packagetype": "sdist",
"python_version": "source",
"requires_python": "<3.12,>=3.8",
"size": 141451,
"upload_time": "2024-07-01T21:31:13",
"upload_time_iso_8601": "2024-07-01T21:31:13.744563Z",
"url": "https://files.pythonhosted.org/packages/a7/92/1cae19960134df7d1058bfdcc98906971ecba6b712933c8c4191725d58bd/rapid_qc_ms-1.1.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-07-01 21:31:13",
"github": false,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"lcname": "rapid-qc-ms"
}