| Name | rdeditor JSON |
| Version |
0.5.0.1
 JSON
JSON |
| download |
| home_page | None |
| Summary | An RDKit based molecule editor using PySide |
| upload_time | 2024-09-17 07:55:02 |
| maintainer | None |
| docs_url | None |
| author | None |
| requires_python | >=3.10 |
| license | LGPL |
| keywords |
rdkit
molecule
editor
pyside
|
| VCS |
 |
| bugtrack_url |
|
| requirements |
No requirements were recorded.
|
| Travis-CI |
No Travis.
|
| coveralls test coverage |
No coveralls.
|
# rdeditor
Simple RDKit molecule editor GUI using PySide6
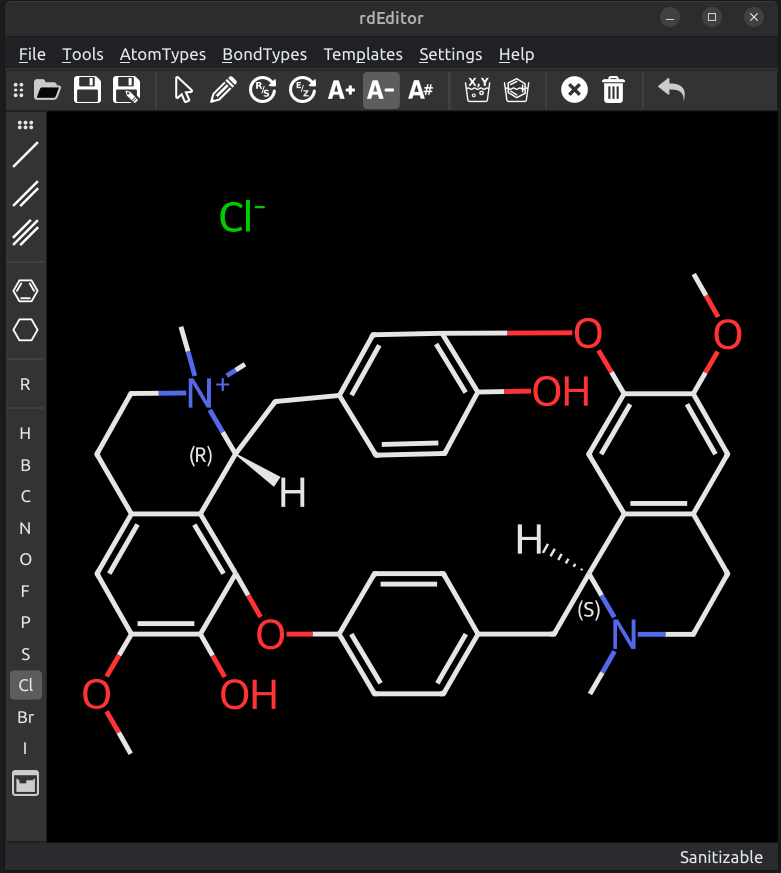
## Installation
- requirements
RDKit, NumPy, PySide6 and pyqtdarktheme should be automatically installed when installed via pip.
- installation
```bash
pip install rdeditor
```
A launch script will also be added so that it can be started from the command line via the `rdEditor` command.
## Usage
Can be started from the command line with `rdEditor` or `rdEditor your_molecule.mol` to start edit an existing molecule.
Interactions with the molecule are done via clicking and dragging on the canvas, atoms or bonds. A choice of tools is available.
To edit a molecule, select the pen tool, and an atom, bond or template type and click on the canvas to add it.
Clicking:
- When clicking existing atoms or bonds, the clicked atom or bond will be modified, depending on the atom or bond type selected.
- If a bondtype is selected, the bond will be added to the clicked atom with a carbon atom at the other end.
- If a bondtype is selected and a bond is clicked, the bond will be changed to that type if different.
- If you click multiple times on a bond with an atomtype selected, the bondtype will cycle between single, double and triple bond.
Dragging:
- If you click and drag from an atom, the selected atomtype will be added at the end of a single bond.
- Dragging between two atoms will add a bond between them. If a bondtype is selected, the bondorder will correspond to the bondtype selected.
- Dragging on the canvas with an atomtype will add two atoms of that type with a single bond between them.
- Dragging on the canvas with a bondtype will simply add that bond with carbon atoms at both ends.
Templates:
- Templates work kind of like atoms, so if you click on an atom, the template will be added directly to that atom.
- If a template is selected and dragged from an atom, the template will be added with a single bond to the clicked atom.
- Some templates can also be added to bonds by clicking on the middle of the bond.
- Dragging on a canvas with a template will add a carbon atom and a single bond to the template.
Other actions:
- most other actions (R/S, E/Z, Increase/Decrease charge, Adjust atom number) works by clikcing on existing atoms.
#### Top Menu:

From left to right
- Open: Open a molfile
- Save: Save current molecule
- Save As: Save current molecule with a new name
- Arrow: Select tool. Click on an atom to select it, click on the canvas to deselect. Clicking on multiple atoms one after another will select them, but only the lastly clicked one will be highlighted in red and used for operations, such as bond creation to another existing atom.
- Pen: Add tool. Clicking on an existing atom will add the current selected atom type to that atom with a single bond. Clicking on the canvas will add a disconnected atom. Clicking on a bond will cycle through single, double and triple bonds.
- R/S: Change the stereo chemistry of the selected atom
- E/Z: Change E/Z stereo of double bonds
- Increase/Decrease charge: Will increase or decrease the charge of the atom clicked
- Set atommap or R-group number: Will set the atommap or R-group number of the atom clicked
- clean up coordinates: recalculate coordinates disregarding existing coordinates.
- clean up chemistry. Sanitize and/or Kekulize the molecule.
- Delete atom/bond:
- Clear Canvas
- Undo.
#### Side Bar:

Most commonly used bond types, and atom types can be selected. Templates and R-group (dummy atoms) are also accessible. A Periodic table is accessible for exotic atom types.
#### Dropdown menus
Access to all standard operations as well as less used atom types and bond-types.
#### Settings
Themes can be selected from the ones available on your platform (Mac/Linux/Windows).
The debug level can be selected
Cleanup settings can be selected if the molecule should be sanitized or kekulized during cleanup.
## Development
Instructions to set it up in editable modes and instructions for eventual contributions can be found in the [DEVELOPER.md](./DEVELOPER.md) file.
Please reach out first, there may be a relevant development branch.
## Additional Reading
I wrote a blog post with an overview of the structure of the code.
[https://www.wildcardconsulting.dk/rdeditor-an-open-source-molecular-editor-based-using-python-pyside2-and-rdkit/](https://www.wildcardconsulting.dk/rdeditor-an-open-source-molecular-editor-based-using-python-pyside2-and-rdkit/)
We also published a preprint on ChemRxiv: [https://chemrxiv.org/engage/chemrxiv/article-details/65e6dcfa9138d23161b2979c](https://chemrxiv.org/engage/chemrxiv/article-details/65e6dcfa9138d23161b2979c)
## ISSUES
Please report issues at GitHub, it's tough getting all corners of a GUI tested.
Raw data
{
"_id": null,
"home_page": null,
"name": "rdeditor",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": null,
"keywords": "RDKit, molecule, editor, pyside",
"author": null,
"author_email": "Esben Jannik Bjerrum <esbenjannik@rocketmail.com>",
"download_url": "https://files.pythonhosted.org/packages/4c/82/8aafd7d4dfa4fe407914aae800ee1c4488f4c917cb6cbaa8f235aefe7dfe/rdeditor-0.5.0.1.tar.gz",
"platform": null,
"description": "# rdeditor\n\nSimple RDKit molecule editor GUI using PySide6\n\n\n\n## Installation\n\n- requirements\n\nRDKit, NumPy, PySide6 and pyqtdarktheme should be automatically installed when installed via pip.\n\n- installation\n\n```bash\npip install rdeditor\n\n```\n\nA launch script will also be added so that it can be started from the command line via the `rdEditor` command.\n\n## Usage\n\nCan be started from the command line with `rdEditor` or `rdEditor your_molecule.mol` to start edit an existing molecule.\nInteractions with the molecule are done via clicking and dragging on the canvas, atoms or bonds. A choice of tools is available.\n\nTo edit a molecule, select the pen tool, and an atom, bond or template type and click on the canvas to add it.\n\nClicking:\n\n- When clicking existing atoms or bonds, the clicked atom or bond will be modified, depending on the atom or bond type selected.\n- If a bondtype is selected, the bond will be added to the clicked atom with a carbon atom at the other end.\n- If a bondtype is selected and a bond is clicked, the bond will be changed to that type if different.\n- If you click multiple times on a bond with an atomtype selected, the bondtype will cycle between single, double and triple bond.\n\nDragging:\n\n- If you click and drag from an atom, the selected atomtype will be added at the end of a single bond.\n- Dragging between two atoms will add a bond between them. If a bondtype is selected, the bondorder will correspond to the bondtype selected.\n- Dragging on the canvas with an atomtype will add two atoms of that type with a single bond between them.\n- Dragging on the canvas with a bondtype will simply add that bond with carbon atoms at both ends.\n\nTemplates:\n\n- Templates work kind of like atoms, so if you click on an atom, the template will be added directly to that atom.\n- If a template is selected and dragged from an atom, the template will be added with a single bond to the clicked atom.\n- Some templates can also be added to bonds by clicking on the middle of the bond.\n- Dragging on a canvas with a template will add a carbon atom and a single bond to the template.\n\nOther actions:\n\n- most other actions (R/S, E/Z, Increase/Decrease charge, Adjust atom number) works by clikcing on existing atoms.\n\n#### Top Menu:\n\n\n\nFrom left to right\n\n- Open: Open a molfile\n- Save: Save current molecule\n- Save As: Save current molecule with a new name\n\n- Arrow: Select tool. Click on an atom to select it, click on the canvas to deselect. Clicking on multiple atoms one after another will select them, but only the lastly clicked one will be highlighted in red and used for operations, such as bond creation to another existing atom.\n- Pen: Add tool. Clicking on an existing atom will add the current selected atom type to that atom with a single bond. Clicking on the canvas will add a disconnected atom. Clicking on a bond will cycle through single, double and triple bonds.\n- R/S: Change the stereo chemistry of the selected atom\n- E/Z: Change E/Z stereo of double bonds\n- Increase/Decrease charge: Will increase or decrease the charge of the atom clicked\n- Set atommap or R-group number: Will set the atommap or R-group number of the atom clicked\n- clean up coordinates: recalculate coordinates disregarding existing coordinates.\n- clean up chemistry. Sanitize and/or Kekulize the molecule.\n- Delete atom/bond:\n- Clear Canvas\n- Undo.\n\n#### Side Bar:\n\n\n\nMost commonly used bond types, and atom types can be selected. Templates and R-group (dummy atoms) are also accessible. A Periodic table is accessible for exotic atom types.\n\n#### Dropdown menus\n\nAccess to all standard operations as well as less used atom types and bond-types.\n\n#### Settings\n\nThemes can be selected from the ones available on your platform (Mac/Linux/Windows).\n\nThe debug level can be selected\n\nCleanup settings can be selected if the molecule should be sanitized or kekulized during cleanup.\n\n## Development\n\nInstructions to set it up in editable modes and instructions for eventual contributions can be found in the [DEVELOPER.md](./DEVELOPER.md) file.\nPlease reach out first, there may be a relevant development branch.\n\n## Additional Reading\n\nI wrote a blog post with an overview of the structure of the code.\n[https://www.wildcardconsulting.dk/rdeditor-an-open-source-molecular-editor-based-using-python-pyside2-and-rdkit/](https://www.wildcardconsulting.dk/rdeditor-an-open-source-molecular-editor-based-using-python-pyside2-and-rdkit/)\n\nWe also published a preprint on ChemRxiv: [https://chemrxiv.org/engage/chemrxiv/article-details/65e6dcfa9138d23161b2979c](https://chemrxiv.org/engage/chemrxiv/article-details/65e6dcfa9138d23161b2979c)\n\n## ISSUES\n\nPlease report issues at GitHub, it's tough getting all corners of a GUI tested.\n",
"bugtrack_url": null,
"license": "LGPL",
"summary": "An RDKit based molecule editor using PySide",
"version": "0.5.0.1",
"project_urls": {
"Bug Tracker": "https://github.com/ebjerrum/rdeditor/issues",
"ChemRxiv Preprint": "https://chemrxiv.org/engage/chemrxiv/article-details/65e6dcfa9138d23161b2979c",
"Citation": "https://github.com/ebjerrum/rdeditor/blob/master/CITATION.md",
"Developer Guide": "https://github.com/ebjerrum/rdeditor/blob/master/DEVELOPER.md",
"Documentation": "https://github.com/ebjerrum/rdeditor/blob/master/README.md",
"Homepage": "http://github.com/ebjerrum/rdeditor",
"Source Code": "https://github.com/ebjerrum/rdeditor"
},
"split_keywords": [
"rdkit",
" molecule",
" editor",
" pyside"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "4a32a51523373944c1a825bcd6f779286a4e8a577f8c4f5c6dcea9c38fac6e44",
"md5": "d94f2a996601a84cee600aae0f9502da",
"sha256": "538c7fa6b2fce125ac956c76fe30ac7b6c5910dc3fcb177394ad35ad926f3e65"
},
"downloads": -1,
"filename": "rdeditor-0.5.0.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "d94f2a996601a84cee600aae0f9502da",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10",
"size": 361009,
"upload_time": "2024-09-17T07:55:01",
"upload_time_iso_8601": "2024-09-17T07:55:01.253003Z",
"url": "https://files.pythonhosted.org/packages/4a/32/a51523373944c1a825bcd6f779286a4e8a577f8c4f5c6dcea9c38fac6e44/rdeditor-0.5.0.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "4c828aafd7d4dfa4fe407914aae800ee1c4488f4c917cb6cbaa8f235aefe7dfe",
"md5": "cc7defc424a31831228686a8e11fd167",
"sha256": "6afdcd9bee8ef0d19694113564187b4f7fbc54012c364ff684993e01166f8cd1"
},
"downloads": -1,
"filename": "rdeditor-0.5.0.1.tar.gz",
"has_sig": false,
"md5_digest": "cc7defc424a31831228686a8e11fd167",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 425946,
"upload_time": "2024-09-17T07:55:02",
"upload_time_iso_8601": "2024-09-17T07:55:02.513581Z",
"url": "https://files.pythonhosted.org/packages/4c/82/8aafd7d4dfa4fe407914aae800ee1c4488f4c917cb6cbaa8f235aefe7dfe/rdeditor-0.5.0.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-09-17 07:55:02",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "ebjerrum",
"github_project": "rdeditor",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "rdeditor"
}
