[](https://github.com/Armavica/rebop/actions/)
[](https://crates.io/crates/rebop/)
[](https://docs.rs/rebop/)
# rebop
rebop is a fast stochastic simulator for well-mixed chemical
reaction networks.
Performance and ergonomics are taken very seriously. For this reason,
two independent APIs are provided to describe and simulate reaction
networks:
* a macro-based DSL implemented by [`define_system`], usually the
most efficient, but that requires to compile a rust program;
* a function-based API implemented by the module [`gillespie`], also
available through Python bindings. This one does not require a rust
compilation and allows the system to be defined at run time. It is
typically 2 or 3 times slower than the macro DSL, but still faster
than all other software tried.
## The macro DSL
It currently only supports reaction rates defined by the law of mass
action. The following macro defines a dimerization reaction network
naturally:
```rust
use rebop::define_system;
define_system! {
r_tx r_tl r_dim r_decay_mRNA r_decay_prot;
Dimers { gene, mRNA, protein, dimer }
transcription : gene => gene + mRNA @ r_tx
translation : mRNA => mRNA + protein @ r_tl
dimerization : 2 protein => dimer @ r_dim
decay_mRNA : mRNA => @ r_decay_mRNA
decay_protein : protein => @ r_decay_prot
}
```
To simulate the system, put this definition in a rust code file and
instantiate the problem, set the parameters, the initial values, and
launch the simulation:
```rust
let mut problem = Dimers::new();
problem.r_tx = 25.0;
problem.r_tl = 1000.0;
problem.r_dim = 0.001;
problem.r_decay_mRNA = 0.1;
problem.r_decay_prot = 1.0;
problem.gene = 1;
problem.advance_until(1.0);
println!("t = {}: dimer = {}", problem.t, problem.dimer);
```
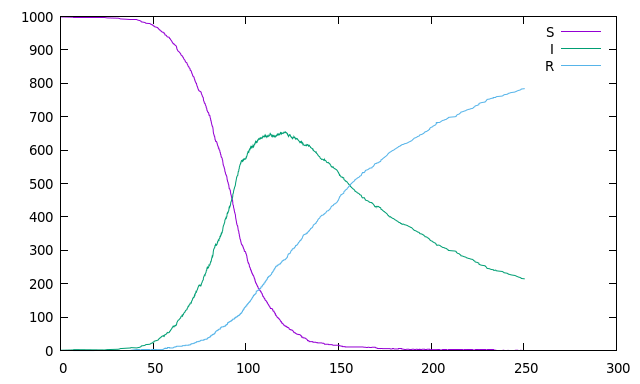
Or for the classic SIR example:
```rust
use rebop::define_system;
define_system! {
r_inf r_heal;
SIR { S, I, R }
infection : S + I => 2 I @ r_inf
healing : I => R @ r_heal
}
fn main() {
let mut problem = SIR::new();
problem.r_inf = 1e-4;
problem.r_heal = 0.01;
problem.S = 999;
problem.I = 1;
println!("time,S,I,R");
for t in 0..250 {
problem.advance_until(t as f64);
println!("{},{},{},{}", problem.t, problem.S, problem.I, problem.R);
}
}
```
which can produce an output similar to this one:
## Python bindings
This API shines through the Python bindings which allow one to
define a model easily:
```python
import rebop
sir = rebop.Gillespie()
sir.add_reaction(1e-4, ['S', 'I'], ['I', 'I'])
sir.add_reaction(0.01, ['I'], ['R'])
print(sir)
ds = sir.run({'S': 999, 'I': 1}, tmax=250, nb_steps=250)
```
You can test this code by installing `rebop` from PyPI with
`pip install rebop`. To build the Python bindings from source,
the simplest is to clone this git repository and use `maturin
develop`.
## The traditional API
The function-based API underlying the Python package is also available
from Rust, if you want to be able to define models at run time (instead
of at compilation time with the macro DSL demonstrated above).
The SIR model is defined as:
```rust
use rebop::gillespie::{Gillespie, Rate};
let mut sir = Gillespie::new([999, 1, 0]);
// [ S, I, R]
// S + I => 2 I with rate 1e-4
sir.add_reaction(Rate::lma(1e-4, [1, 1, 0]), [-1, 1, 0]);
// I => R with rate 0.01
sir.add_reaction(Rate::lma(0.01, [0, 1, 0]), [0, -1, 1]);
println!("time,S,I,R");
for t in 0..250 {
sir.advance_until(t as f64);
println!("{},{},{},{}", sir.get_time(), sir.get_species(0), sir.get_species(1), sir.get_species(2));
}
```
## Performance
Performance is taken very seriously, and as a result, rebop
outperforms every other package and programming language that we
tried.
*Disclaimer*: Most of this software currently contains much more
features than rebop (e.g. spatial models, custom reaction rates,
etc.). Some of these features might have required them to make
compromises on speed. Moreover, as much as we tried to keep the
comparison fair, some return too much or too little data, or write
them on disk. The baseline that we tried to approach for all these
programs is the following: *the model was just modified, we want
to simulate it `N` times and print regularly spaced measurement
points*. This means that we always include initialization or
(re-)compilation time if applicable. We think that it is the most
typical use-case of a researcher who works on the model. This
benchmark methods allows to record both the initialization time
(y-intercept) and the simulation time per simulation (slope).
Many small benchmarks on toy examples are tracked to guide the
development. To compare the performance with other software,
we used a real-world model of low-medium size (9 species and 16
reactions): the Vilar oscillator (*Mechanisms of noise-resistance
in genetic oscillators*, Vilar et al., PNAS 2002). Here, we
simulate this model from `t=0` to `t=200`, reporting the state at
time intervals of `1` time unit.

We can see that rebop's macro DSL is the fastest of all, both in
time per simulation, and with compilation time included. The second
fastest is rebop's traditional API invoked by convenience through
the Python bindings.
## Features to come
* compartment volumes
* arbitrary reaction rates
* other SSA algorithms
* tau-leaping
* adaptive tau-leaping
* hybrid models (continuous and discrete)
* SBML
* CLI interface
* parameter estimation
* local sensitivity analysis
* parallelization
## Features probably not to come
* events
* space (reaction-diffusion systems)
* rule modelling
## Benchmark ideas
* DSMTS
* purely decoupled exponentials
* ring
* Toggle switch
* LacZ, LacY/LacZ (from STOCKS)
* Lotka Volterra, Michaelis--Menten, Network (from StochSim)
* G protein (from SimBiology)
* Brusselator / Oregonator (from Cellware)
* GAL, repressilator (from Dizzy)
## Similar software
### Maintained
* [GillesPy2](https://github.com/StochSS/GillesPy2)
* [STEPS](https://github.com/CNS-OIST/STEPS)
* [SimBiology](https://fr.mathworks.com/help/simbio/)
* [Copasi](http://copasi.org/)
* [BioNetGen](http://bionetgen.org/)
* [VCell](http://vcell.org/)
* [Smoldyn](http://www.smoldyn.org/)
* [KaSim](https://kappalanguage.org/)
* [StochPy](https://github.com/SystemsBioinformatics/stochpy)
* [BioSimulator.jl](https://github.com/alanderos91/BioSimulator.jl)
* [DiffEqJump.jl](https://github.com/SciML/DiffEqJump.jl)
* [Gillespie.jl](https://github.com/sdwfrost/Gillespie.jl)
* [GillespieSSA2](https://github.com/rcannood/GillespieSSA2)
* [Cayenne](https://github.com/quantumbrake/cayenne)
### Seem unmaintained
* [Dizzy](http://magnet.systemsbiology.net/software/Dizzy/)
* [Cellware](http://www.bii.a-star.edu.sg/achievements/applications/cellware/)
* [STOCKS](https://doi.org/10.1093/bioinformatics/18.3.470)
* [StochSim](http://lenoverelab.org/perso/lenov/stochsim.html)
* [Systems biology toolbox](http://www.sbtoolbox.org/)
* [StochKit](https://github.com/StochSS/StochKit) (successor: GillesPy2)
* [SmartCell](http://software.crg.es/smartcell/)
* [NFsim](http://michaelsneddon.net/nfsim/)
License: MIT
Raw data
{
"_id": null,
"home_page": "https://armavica.github.io/rebop/",
"name": "rebop",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": null,
"keywords": "gillespie-algorithm, systems-biology, stochastic, scientific-computing",
"author": "Virgile Andreani <armavica@ulminfo.fr>",
"author_email": "Virgile Andreani <armavica@ulminfo.fr>",
"download_url": "https://files.pythonhosted.org/packages/3e/2a/02c523a894dc9d5a20df4bc225610b56e0f8e3ab717b03f2421d379d1a75/rebop-0.8.3.tar.gz",
"platform": null,
"description": "[](https://github.com/Armavica/rebop/actions/)\n[](https://crates.io/crates/rebop/)\n[](https://docs.rs/rebop/)\n\n# rebop\n\nrebop is a fast stochastic simulator for well-mixed chemical\nreaction networks.\n\nPerformance and ergonomics are taken very seriously. For this reason,\ntwo independent APIs are provided to describe and simulate reaction\nnetworks:\n\n* a macro-based DSL implemented by [`define_system`], usually the\nmost efficient, but that requires to compile a rust program;\n* a function-based API implemented by the module [`gillespie`], also\navailable through Python bindings. This one does not require a rust\ncompilation and allows the system to be defined at run time. It is\ntypically 2 or 3 times slower than the macro DSL, but still faster\nthan all other software tried.\n\n## The macro DSL\n\nIt currently only supports reaction rates defined by the law of mass\naction. The following macro defines a dimerization reaction network\nnaturally:\n\n```rust\nuse rebop::define_system;\ndefine_system! {\n r_tx r_tl r_dim r_decay_mRNA r_decay_prot;\n Dimers { gene, mRNA, protein, dimer }\n transcription : gene => gene + mRNA @ r_tx\n translation : mRNA => mRNA + protein @ r_tl\n dimerization : 2 protein => dimer @ r_dim\n decay_mRNA : mRNA => @ r_decay_mRNA\n decay_protein : protein => @ r_decay_prot\n}\n```\n\nTo simulate the system, put this definition in a rust code file and\ninstantiate the problem, set the parameters, the initial values, and\nlaunch the simulation:\n\n```rust\nlet mut problem = Dimers::new();\nproblem.r_tx = 25.0;\nproblem.r_tl = 1000.0;\nproblem.r_dim = 0.001;\nproblem.r_decay_mRNA = 0.1;\nproblem.r_decay_prot = 1.0;\nproblem.gene = 1;\nproblem.advance_until(1.0);\nprintln!(\"t = {}: dimer = {}\", problem.t, problem.dimer);\n```\n\nOr for the classic SIR example:\n\n```rust\nuse rebop::define_system;\n\ndefine_system! {\n r_inf r_heal;\n SIR { S, I, R }\n infection : S + I => 2 I @ r_inf\n healing : I => R @ r_heal\n}\n\nfn main() {\n let mut problem = SIR::new();\n problem.r_inf = 1e-4;\n problem.r_heal = 0.01;\n problem.S = 999;\n problem.I = 1;\n println!(\"time,S,I,R\");\n for t in 0..250 {\n problem.advance_until(t as f64);\n println!(\"{},{},{},{}\", problem.t, problem.S, problem.I, problem.R);\n }\n}\n```\n\nwhich can produce an output similar to this one:\n\n\n\n## Python bindings\n\nThis API shines through the Python bindings which allow one to\ndefine a model easily:\n\n```python\nimport rebop\n\nsir = rebop.Gillespie()\nsir.add_reaction(1e-4, ['S', 'I'], ['I', 'I'])\nsir.add_reaction(0.01, ['I'], ['R'])\nprint(sir)\n\nds = sir.run({'S': 999, 'I': 1}, tmax=250, nb_steps=250)\n```\n\nYou can test this code by installing `rebop` from PyPI with\n`pip install rebop`. To build the Python bindings from source,\nthe simplest is to clone this git repository and use `maturin\ndevelop`.\n\n## The traditional API\n\nThe function-based API underlying the Python package is also available\nfrom Rust, if you want to be able to define models at run time (instead\nof at compilation time with the macro DSL demonstrated above).\nThe SIR model is defined as:\n\n```rust\nuse rebop::gillespie::{Gillespie, Rate};\n\nlet mut sir = Gillespie::new([999, 1, 0]);\n// [ S, I, R]\n// S + I => 2 I with rate 1e-4\nsir.add_reaction(Rate::lma(1e-4, [1, 1, 0]), [-1, 1, 0]);\n// I => R with rate 0.01\nsir.add_reaction(Rate::lma(0.01, [0, 1, 0]), [0, -1, 1]);\n\nprintln!(\"time,S,I,R\");\nfor t in 0..250 {\n sir.advance_until(t as f64);\n println!(\"{},{},{},{}\", sir.get_time(), sir.get_species(0), sir.get_species(1), sir.get_species(2));\n}\n```\n\n## Performance\n\nPerformance is taken very seriously, and as a result, rebop\noutperforms every other package and programming language that we\ntried.\n\n*Disclaimer*: Most of this software currently contains much more\nfeatures than rebop (e.g. spatial models, custom reaction rates,\netc.). Some of these features might have required them to make\ncompromises on speed. Moreover, as much as we tried to keep the\ncomparison fair, some return too much or too little data, or write\nthem on disk. The baseline that we tried to approach for all these\nprograms is the following: *the model was just modified, we want\nto simulate it `N` times and print regularly spaced measurement\npoints*. This means that we always include initialization or\n(re-)compilation time if applicable. We think that it is the most\ntypical use-case of a researcher who works on the model. This\nbenchmark methods allows to record both the initialization time\n(y-intercept) and the simulation time per simulation (slope).\n\nMany small benchmarks on toy examples are tracked to guide the\ndevelopment. To compare the performance with other software,\nwe used a real-world model of low-medium size (9 species and 16\nreactions): the Vilar oscillator (*Mechanisms of noise-resistance\nin genetic oscillators*, Vilar et al., PNAS 2002). Here, we\nsimulate this model from `t=0` to `t=200`, reporting the state at\ntime intervals of `1` time unit.\n\n\n\nWe can see that rebop's macro DSL is the fastest of all, both in\ntime per simulation, and with compilation time included. The second\nfastest is rebop's traditional API invoked by convenience through\nthe Python bindings.\n\n## Features to come\n\n* compartment volumes\n* arbitrary reaction rates\n* other SSA algorithms\n* tau-leaping\n* adaptive tau-leaping\n* hybrid models (continuous and discrete)\n* SBML\n* CLI interface\n* parameter estimation\n* local sensitivity analysis\n* parallelization\n\n## Features probably not to come\n\n* events\n* space (reaction-diffusion systems)\n* rule modelling\n\n## Benchmark ideas\n\n* DSMTS\n* purely decoupled exponentials\n* ring\n* Toggle switch\n* LacZ, LacY/LacZ (from STOCKS)\n* Lotka Volterra, Michaelis--Menten, Network (from StochSim)\n* G protein (from SimBiology)\n* Brusselator / Oregonator (from Cellware)\n* GAL, repressilator (from Dizzy)\n\n## Similar software\n\n### Maintained\n\n* [GillesPy2](https://github.com/StochSS/GillesPy2)\n* [STEPS](https://github.com/CNS-OIST/STEPS)\n* [SimBiology](https://fr.mathworks.com/help/simbio/)\n* [Copasi](http://copasi.org/)\n* [BioNetGen](http://bionetgen.org/)\n* [VCell](http://vcell.org/)\n* [Smoldyn](http://www.smoldyn.org/)\n* [KaSim](https://kappalanguage.org/)\n* [StochPy](https://github.com/SystemsBioinformatics/stochpy)\n* [BioSimulator.jl](https://github.com/alanderos91/BioSimulator.jl)\n* [DiffEqJump.jl](https://github.com/SciML/DiffEqJump.jl)\n* [Gillespie.jl](https://github.com/sdwfrost/Gillespie.jl)\n* [GillespieSSA2](https://github.com/rcannood/GillespieSSA2)\n* [Cayenne](https://github.com/quantumbrake/cayenne)\n\n### Seem unmaintained\n\n* [Dizzy](http://magnet.systemsbiology.net/software/Dizzy/)\n* [Cellware](http://www.bii.a-star.edu.sg/achievements/applications/cellware/)\n* [STOCKS](https://doi.org/10.1093/bioinformatics/18.3.470)\n* [StochSim](http://lenoverelab.org/perso/lenov/stochsim.html)\n* [Systems biology toolbox](http://www.sbtoolbox.org/)\n* [StochKit](https://github.com/StochSS/StochKit) (successor: GillesPy2)\n* [SmartCell](http://software.crg.es/smartcell/)\n* [NFsim](http://michaelsneddon.net/nfsim/)\n\nLicense: MIT\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A fast stochastic simulator for chemical reaction networks",
"version": "0.8.3",
"project_urls": {
"Homepage": "https://armavica.github.io/rebop/",
"Source Code": "https://github.com/Armavica/rebop/"
},
"split_keywords": [
"gillespie-algorithm",
" systems-biology",
" stochastic",
" scientific-computing"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "e6dcffe0f6525c37d84cec4ded1afc72e229d11df85c0ee631481ea8bdeb8be5",
"md5": "95b347308d0382b860b317de21c16b39",
"sha256": "62eab853047d9f02ac1c9c76c78daf1af87f0a34348eb2b8308ac7da8935c184"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-macosx_11_0_arm64.whl",
"has_sig": false,
"md5_digest": "95b347308d0382b860b317de21c16b39",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 236818,
"upload_time": "2024-07-18T02:08:22",
"upload_time_iso_8601": "2024-07-18T02:08:22.036698Z",
"url": "https://files.pythonhosted.org/packages/e6/dc/ffe0f6525c37d84cec4ded1afc72e229d11df85c0ee631481ea8bdeb8be5/rebop-0.8.3-cp310-cp310-macosx_11_0_arm64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "a3f00125c602777f47bb0b8b81f5070b8f6fea96c4a81852a6fe29e319a16068",
"md5": "6126723215e13778d397f848ec8ef345",
"sha256": "067315e383202af787b44c48065cabb5bc13df55209455237c31dace658b435d"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"has_sig": false,
"md5_digest": "6126723215e13778d397f848ec8ef345",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 272971,
"upload_time": "2024-07-18T02:07:47",
"upload_time_iso_8601": "2024-07-18T02:07:47.484027Z",
"url": "https://files.pythonhosted.org/packages/a3/f0/0125c602777f47bb0b8b81f5070b8f6fea96c4a81852a6fe29e319a16068/rebop-0.8.3-cp310-cp310-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "4631820aa4eb59283ae7575d86342669818595203e178ccc55d85337a589fc81",
"md5": "3ecf5e6621c183aa4640d41a9a8b207b",
"sha256": "d6efc7034b9123fb6e86e556cf350299885828086d5c08af5a4391a2aedf4893"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"has_sig": false,
"md5_digest": "3ecf5e6621c183aa4640d41a9a8b207b",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 280374,
"upload_time": "2024-07-18T02:07:53",
"upload_time_iso_8601": "2024-07-18T02:07:53.242707Z",
"url": "https://files.pythonhosted.org/packages/46/31/820aa4eb59283ae7575d86342669818595203e178ccc55d85337a589fc81/rebop-0.8.3-cp310-cp310-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "187da864d104e1b9097bdd3c8d182673ae9a015633303da21e56455d9dd0db7e",
"md5": "accb04992d04f4a56f61b67acb0bcc53",
"sha256": "2ab92a47a06524ca05799175869a52a02a71fa5ae14d13786cc269dfbb3759fb"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"has_sig": false,
"md5_digest": "accb04992d04f4a56f61b67acb0bcc53",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 294568,
"upload_time": "2024-07-18T02:07:59",
"upload_time_iso_8601": "2024-07-18T02:07:59.770776Z",
"url": "https://files.pythonhosted.org/packages/18/7d/a864d104e1b9097bdd3c8d182673ae9a015633303da21e56455d9dd0db7e/rebop-0.8.3-cp310-cp310-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "31f4c3441c45459059486774d296baf4a8859ace3e5b6b9dd54d1da290484361",
"md5": "19cae96ac9cdc8b454c3799beb4ec021",
"sha256": "0a6310bbba871dc6855b183570258870ac58b552662c83287d6fe33ee222a7fb"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"has_sig": false,
"md5_digest": "19cae96ac9cdc8b454c3799beb4ec021",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 352549,
"upload_time": "2024-07-18T02:08:05",
"upload_time_iso_8601": "2024-07-18T02:08:05.599570Z",
"url": "https://files.pythonhosted.org/packages/31/f4/c3441c45459059486774d296baf4a8859ace3e5b6b9dd54d1da290484361/rebop-0.8.3-cp310-cp310-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "9f66628458b54f8e40ce1e4af905fdee7a0178a13dec79b2215de649b5bbaf92",
"md5": "56eb163271ff713a6378f5ec6dda494d",
"sha256": "9467f55e348903e9072cbb336b43d8ba1b5cea21cffb6a7bd755785afacd61c1"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"has_sig": false,
"md5_digest": "56eb163271ff713a6378f5ec6dda494d",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 267345,
"upload_time": "2024-07-18T02:08:16",
"upload_time_iso_8601": "2024-07-18T02:08:16.401385Z",
"url": "https://files.pythonhosted.org/packages/9f/66/628458b54f8e40ce1e4af905fdee7a0178a13dec79b2215de649b5bbaf92/rebop-0.8.3-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "2571107f069dba6d8d3a4ea5bbd6886011fa8d7d2cd0d928c2a79ec8a26fe041",
"md5": "0ec1a941b474a63e729879bd903adc75",
"sha256": "ac6e09ec6ce6bef5b82b39f4089aa2d5c502bc538015c41e447fdb082a2db7da"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.whl",
"has_sig": false,
"md5_digest": "0ec1a941b474a63e729879bd903adc75",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 276016,
"upload_time": "2024-07-18T02:08:11",
"upload_time_iso_8601": "2024-07-18T02:08:11.501933Z",
"url": "https://files.pythonhosted.org/packages/25/71/107f069dba6d8d3a4ea5bbd6886011fa8d7d2cd0d928c2a79ec8a26fe041/rebop-0.8.3-cp310-cp310-manylinux_2_5_i686.manylinux1_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "7faab1d21b67208122cf937da72691b16626f93d01ecb50d9a3e947205ac244b",
"md5": "09a527ac8cbc49b25f909347120b7b5a",
"sha256": "2d3367fd93f0a72a21d33e516546863842eabadb7b6dfff030888629b195f51b"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-musllinux_1_2_aarch64.whl",
"has_sig": false,
"md5_digest": "09a527ac8cbc49b25f909347120b7b5a",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 452388,
"upload_time": "2024-07-18T02:08:28",
"upload_time_iso_8601": "2024-07-18T02:08:28.851471Z",
"url": "https://files.pythonhosted.org/packages/7f/aa/b1d21b67208122cf937da72691b16626f93d01ecb50d9a3e947205ac244b/rebop-0.8.3-cp310-cp310-musllinux_1_2_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "727730d3c6793474876afcd7b47854c604277facc6ee79edef3157ebc1583103",
"md5": "9ef6a8dab772dcc374d771585360cdf7",
"sha256": "2f709ac452be09fd674c8b3532552841ea64d7ac504b07b6e850c98d65602484"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-musllinux_1_2_armv7l.whl",
"has_sig": false,
"md5_digest": "9ef6a8dab772dcc374d771585360cdf7",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 542266,
"upload_time": "2024-07-18T02:08:35",
"upload_time_iso_8601": "2024-07-18T02:08:35.282169Z",
"url": "https://files.pythonhosted.org/packages/72/77/30d3c6793474876afcd7b47854c604277facc6ee79edef3157ebc1583103/rebop-0.8.3-cp310-cp310-musllinux_1_2_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "1d089eeb0df56d0f54a7ea9aec967a3cc7135c4448a18ce450af213710fbbcc7",
"md5": "0b3de9b2c034622cf4135286e4bb89e4",
"sha256": "c45edd5db3d3bae02dcce0aa48a175244248722d2ea1ed11e617c04199a946c5"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-musllinux_1_2_i686.whl",
"has_sig": false,
"md5_digest": "0b3de9b2c034622cf4135286e4bb89e4",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 456166,
"upload_time": "2024-07-18T02:08:41",
"upload_time_iso_8601": "2024-07-18T02:08:41.089059Z",
"url": "https://files.pythonhosted.org/packages/1d/08/9eeb0df56d0f54a7ea9aec967a3cc7135c4448a18ce450af213710fbbcc7/rebop-0.8.3-cp310-cp310-musllinux_1_2_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "b72b0bee60ecfc19490fa3875cae91d0955187c52f523464e15fbcd8c2686c74",
"md5": "7517b8c974b387cc5b243f062f606f0f",
"sha256": "6537281dd053ae7b50523f1c0f36ea3ca9968e3c9ad9c71c57424d9a7ad4a9b9"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-cp310-musllinux_1_2_x86_64.whl",
"has_sig": false,
"md5_digest": "7517b8c974b387cc5b243f062f606f0f",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 437999,
"upload_time": "2024-07-18T02:08:46",
"upload_time_iso_8601": "2024-07-18T02:08:46.699125Z",
"url": "https://files.pythonhosted.org/packages/b7/2b/0bee60ecfc19490fa3875cae91d0955187c52f523464e15fbcd8c2686c74/rebop-0.8.3-cp310-cp310-musllinux_1_2_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "3af589000c8022e7f2aae72f923db892cb4ade3e8b233be69fb1ac591cba202a",
"md5": "ea4b515dabe7263ce7806ee039407c8c",
"sha256": "e4cd907b70f4d5e2b7b917d2d5ae0ea03f08e48f71e2e81aba5ff422f0efc46b"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-none-win32.whl",
"has_sig": false,
"md5_digest": "ea4b515dabe7263ce7806ee039407c8c",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 147597,
"upload_time": "2024-07-18T02:08:56",
"upload_time_iso_8601": "2024-07-18T02:08:56.322162Z",
"url": "https://files.pythonhosted.org/packages/3a/f5/89000c8022e7f2aae72f923db892cb4ade3e8b233be69fb1ac591cba202a/rebop-0.8.3-cp310-none-win32.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "337baef4b857662f54c2c2b82867ff7ceeb054c301a5f3e53821db3756f70e48",
"md5": "390740e0399d3c188c619e2111f0d372",
"sha256": "c168d19a0066ad009d9a53bd6e87ffef4a3e468bf909602abd94704394a962aa"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp310-none-win_amd64.whl",
"has_sig": false,
"md5_digest": "390740e0399d3c188c619e2111f0d372",
"packagetype": "bdist_wheel",
"python_version": "cp310",
"requires_python": ">=3.10",
"size": 153147,
"upload_time": "2024-07-18T02:08:52",
"upload_time_iso_8601": "2024-07-18T02:08:52.914297Z",
"url": "https://files.pythonhosted.org/packages/33/7b/aef4b857662f54c2c2b82867ff7ceeb054c301a5f3e53821db3756f70e48/rebop-0.8.3-cp310-none-win_amd64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "b140824d03311bb24cd56715c5438f31faa8e436ce8331d83cdb6ba6f908adde",
"md5": "421495540c70257580267e81d6b9410c",
"sha256": "4fb8856165ee279621619941de6bff031ea5effa99e4394fee89a820171aef16"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-macosx_10_12_x86_64.whl",
"has_sig": false,
"md5_digest": "421495540c70257580267e81d6b9410c",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 241478,
"upload_time": "2024-07-18T02:08:25",
"upload_time_iso_8601": "2024-07-18T02:08:25.876301Z",
"url": "https://files.pythonhosted.org/packages/b1/40/824d03311bb24cd56715c5438f31faa8e436ce8331d83cdb6ba6f908adde/rebop-0.8.3-cp311-cp311-macosx_10_12_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "9d2cc1917856e4ee36a48da7a227ee6c1bd93b28e15ff4a0298c928bcdc212bf",
"md5": "4f3d8ccceb454503de041e4688335451",
"sha256": "38ab02e4898eff13e9a9ca02069c21b89d64652444140d274ec6cfa0c1686624"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-macosx_11_0_arm64.whl",
"has_sig": false,
"md5_digest": "4f3d8ccceb454503de041e4688335451",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 236564,
"upload_time": "2024-07-18T02:08:23",
"upload_time_iso_8601": "2024-07-18T02:08:23.083204Z",
"url": "https://files.pythonhosted.org/packages/9d/2c/c1917856e4ee36a48da7a227ee6c1bd93b28e15ff4a0298c928bcdc212bf/rebop-0.8.3-cp311-cp311-macosx_11_0_arm64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "ac0061813915b373bbb269b20b71c3a8a78f062124928a541b2c7573519fddd5",
"md5": "c145df3ce51f4545c9eeca51c4729fca",
"sha256": "2cc3d02fba75e8fcd6a7ca78d7b2ed460ed5f2125edb1355d8b7c87303cbde7e"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"has_sig": false,
"md5_digest": "c145df3ce51f4545c9eeca51c4729fca",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 272321,
"upload_time": "2024-07-18T02:07:49",
"upload_time_iso_8601": "2024-07-18T02:07:49.088294Z",
"url": "https://files.pythonhosted.org/packages/ac/00/61813915b373bbb269b20b71c3a8a78f062124928a541b2c7573519fddd5/rebop-0.8.3-cp311-cp311-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "fb64a9409349408925ce0817000cf46e79684fa068951a7c3ed8fd02a006ff16",
"md5": "46ec747a2c1f8173371cecf97a87f16a",
"sha256": "ffa6209e7149082412895c11a0ed9f4735de92763757609c6e44efa3047037ac"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"has_sig": false,
"md5_digest": "46ec747a2c1f8173371cecf97a87f16a",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 280060,
"upload_time": "2024-07-18T02:07:55",
"upload_time_iso_8601": "2024-07-18T02:07:55.020925Z",
"url": "https://files.pythonhosted.org/packages/fb/64/a9409349408925ce0817000cf46e79684fa068951a7c3ed8fd02a006ff16/rebop-0.8.3-cp311-cp311-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "b6f0dc7ed13179f53a5a997c5aa3a5c1a6c0f23f83460673e871985858951702",
"md5": "3d31942dad58324d1cf5c250f0516ed2",
"sha256": "f24e73f511184d699d6c007af2b4b68946ad3a348cc9d95af4d9112241726fbd"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"has_sig": false,
"md5_digest": "3d31942dad58324d1cf5c250f0516ed2",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 294322,
"upload_time": "2024-07-18T02:08:01",
"upload_time_iso_8601": "2024-07-18T02:08:01.183673Z",
"url": "https://files.pythonhosted.org/packages/b6/f0/dc7ed13179f53a5a997c5aa3a5c1a6c0f23f83460673e871985858951702/rebop-0.8.3-cp311-cp311-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "fa30298fbcf5e9c39dc624ce7f99ce98ecc4d017dd1ad1b8628ccaa9b76fc48b",
"md5": "729654399635fd230d90b4a191c50108",
"sha256": "a09f376100412fc5329b76e82cf9b0a2cfe1a6f1155b8660ec2e6e139c71a829"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"has_sig": false,
"md5_digest": "729654399635fd230d90b4a191c50108",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 353099,
"upload_time": "2024-07-18T02:08:07",
"upload_time_iso_8601": "2024-07-18T02:08:07.224297Z",
"url": "https://files.pythonhosted.org/packages/fa/30/298fbcf5e9c39dc624ce7f99ce98ecc4d017dd1ad1b8628ccaa9b76fc48b/rebop-0.8.3-cp311-cp311-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "9c2ad4525bd1b245614dcc508d42731805124bbb5988398a3376c0ab2637252f",
"md5": "738d3066536fed7fecc9410e26cdcba8",
"sha256": "bdb24a38c3b9f660817e02bf0904a29dd76b19e4622671cd3e05961396976536"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"has_sig": false,
"md5_digest": "738d3066536fed7fecc9410e26cdcba8",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 267115,
"upload_time": "2024-07-18T02:08:17",
"upload_time_iso_8601": "2024-07-18T02:08:17.710713Z",
"url": "https://files.pythonhosted.org/packages/9c/2a/d4525bd1b245614dcc508d42731805124bbb5988398a3376c0ab2637252f/rebop-0.8.3-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "e585c83517f99d7972a1eef539db816df578957a0263c7a4ef64507e7067d3ad",
"md5": "f1093dfa6a86ff017a62d89179cb686f",
"sha256": "601074463a05411b7d75ab025fc107dba0c978c0fe5d86d93acecf9af957e44b"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.whl",
"has_sig": false,
"md5_digest": "f1093dfa6a86ff017a62d89179cb686f",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 275695,
"upload_time": "2024-07-18T02:08:12",
"upload_time_iso_8601": "2024-07-18T02:08:12.941952Z",
"url": "https://files.pythonhosted.org/packages/e5/85/c83517f99d7972a1eef539db816df578957a0263c7a4ef64507e7067d3ad/rebop-0.8.3-cp311-cp311-manylinux_2_5_i686.manylinux1_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "03e6bec563fffa528f8492a6ed8c277414f8d96751e5f519d559d7702b1dcf2a",
"md5": "8800879e1fd881086b9747b76dd2d587",
"sha256": "9d653a5c45879e4b07f2c00aeab280ffc77123959ee30a1cb0f728d29a7500ad"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-musllinux_1_2_aarch64.whl",
"has_sig": false,
"md5_digest": "8800879e1fd881086b9747b76dd2d587",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 451959,
"upload_time": "2024-07-18T02:08:30",
"upload_time_iso_8601": "2024-07-18T02:08:30.465043Z",
"url": "https://files.pythonhosted.org/packages/03/e6/bec563fffa528f8492a6ed8c277414f8d96751e5f519d559d7702b1dcf2a/rebop-0.8.3-cp311-cp311-musllinux_1_2_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "65feb01cc42a37bd8ca6364474e23b0e408f4ae96e345a1a788d3b2413af9262",
"md5": "b842b8f94fca49e9516d7d0450e6b352",
"sha256": "dd30730f67518868cba12dc029357492813b9f7bdf0606b329d1e6deca8c2adb"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-musllinux_1_2_armv7l.whl",
"has_sig": false,
"md5_digest": "b842b8f94fca49e9516d7d0450e6b352",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 541779,
"upload_time": "2024-07-18T02:08:36",
"upload_time_iso_8601": "2024-07-18T02:08:36.666424Z",
"url": "https://files.pythonhosted.org/packages/65/fe/b01cc42a37bd8ca6364474e23b0e408f4ae96e345a1a788d3b2413af9262/rebop-0.8.3-cp311-cp311-musllinux_1_2_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "4000730bbeb37e7eadc57f73d10a196a1bd119b538792b5c08f15ebc80a6d516",
"md5": "f37b53e42146a5393eb4c513bfc2040e",
"sha256": "ef8d7623e62223f2fadb1fbf78c285bf0006bae58a5dbef6c0e4986939d87fdb"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-musllinux_1_2_i686.whl",
"has_sig": false,
"md5_digest": "f37b53e42146a5393eb4c513bfc2040e",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 455889,
"upload_time": "2024-07-18T02:08:42",
"upload_time_iso_8601": "2024-07-18T02:08:42.519444Z",
"url": "https://files.pythonhosted.org/packages/40/00/730bbeb37e7eadc57f73d10a196a1bd119b538792b5c08f15ebc80a6d516/rebop-0.8.3-cp311-cp311-musllinux_1_2_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "4c9f3f1e17db86363fd39ec02ff91711531f0767bcefb5d30d5081c3a8130619",
"md5": "f714effbbd69bead981e2ab41120bdc6",
"sha256": "aa2f1ec9a34e76d53c489048f660d106d2cc1f977b6f2ab15ae19723acfaca84"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-cp311-musllinux_1_2_x86_64.whl",
"has_sig": false,
"md5_digest": "f714effbbd69bead981e2ab41120bdc6",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 437777,
"upload_time": "2024-07-18T02:08:47",
"upload_time_iso_8601": "2024-07-18T02:08:47.998721Z",
"url": "https://files.pythonhosted.org/packages/4c/9f/3f1e17db86363fd39ec02ff91711531f0767bcefb5d30d5081c3a8130619/rebop-0.8.3-cp311-cp311-musllinux_1_2_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "de4c2aab3358ca84f923fa681528e372c39e9a38ed455a4c6ffb3cb612472913",
"md5": "38c486165b53fa5bf513d75711bae834",
"sha256": "3e0db2a4fcadcfb5284faa2a25d4e854c992f169c88d0990f2207e88a7d03fc4"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-none-win32.whl",
"has_sig": false,
"md5_digest": "38c486165b53fa5bf513d75711bae834",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 147440,
"upload_time": "2024-07-18T02:08:57",
"upload_time_iso_8601": "2024-07-18T02:08:57.438666Z",
"url": "https://files.pythonhosted.org/packages/de/4c/2aab3358ca84f923fa681528e372c39e9a38ed455a4c6ffb3cb612472913/rebop-0.8.3-cp311-none-win32.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "6d1c31aa30995a1623917a16b0205e24a75df19eba8133be2955e759536f2089",
"md5": "3617172d5e5ddf32e5b6e9486d47074d",
"sha256": "9f2c408cd7d204deafc1acaa951865d38e659dd0e8deee290d5c2bf10651cff2"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp311-none-win_amd64.whl",
"has_sig": false,
"md5_digest": "3617172d5e5ddf32e5b6e9486d47074d",
"packagetype": "bdist_wheel",
"python_version": "cp311",
"requires_python": ">=3.10",
"size": 152830,
"upload_time": "2024-07-18T02:08:53",
"upload_time_iso_8601": "2024-07-18T02:08:53.977017Z",
"url": "https://files.pythonhosted.org/packages/6d/1c/31aa30995a1623917a16b0205e24a75df19eba8133be2955e759536f2089/rebop-0.8.3-cp311-none-win_amd64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "7ac3ee9067a08c25ea62e4a854c80d1b9b1367f0de47b58831296e49158dc6c5",
"md5": "8733e4a254f71b0cc25e9cc92c29ba56",
"sha256": "ffee39002d5b92b5a1fdaaefaf38af526f9fa81a6d9c085796212988cac9a8e2"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-macosx_10_12_x86_64.whl",
"has_sig": false,
"md5_digest": "8733e4a254f71b0cc25e9cc92c29ba56",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 241278,
"upload_time": "2024-07-18T02:08:27",
"upload_time_iso_8601": "2024-07-18T02:08:27.628884Z",
"url": "https://files.pythonhosted.org/packages/7a/c3/ee9067a08c25ea62e4a854c80d1b9b1367f0de47b58831296e49158dc6c5/rebop-0.8.3-cp312-cp312-macosx_10_12_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "e169fa65dbebc916fa3b50d392d1b09b0b7730a32aacfc466ae4e241dcf19292",
"md5": "5f0d8f97ca9f86e88e272425c2deb0d5",
"sha256": "4b34401ff14384fa4c15059e358e88d2e977e12646fb4999149522c5bd1fa7fe"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-macosx_11_0_arm64.whl",
"has_sig": false,
"md5_digest": "5f0d8f97ca9f86e88e272425c2deb0d5",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 236188,
"upload_time": "2024-07-18T02:08:24",
"upload_time_iso_8601": "2024-07-18T02:08:24.214925Z",
"url": "https://files.pythonhosted.org/packages/e1/69/fa65dbebc916fa3b50d392d1b09b0b7730a32aacfc466ae4e241dcf19292/rebop-0.8.3-cp312-cp312-macosx_11_0_arm64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "74914560bf922875f6a5b6fb7d86bad1be2894053e3cd61305d7811c5ced33a2",
"md5": "ba16862b8f87312255ffa2e3fcd08039",
"sha256": "e90fcb646ca12365f2c0d0b818a5256acb12c937c4e2161e1b532c9cb461bfdd"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"has_sig": false,
"md5_digest": "ba16862b8f87312255ffa2e3fcd08039",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 272285,
"upload_time": "2024-07-18T02:07:50",
"upload_time_iso_8601": "2024-07-18T02:07:50.579781Z",
"url": "https://files.pythonhosted.org/packages/74/91/4560bf922875f6a5b6fb7d86bad1be2894053e3cd61305d7811c5ced33a2/rebop-0.8.3-cp312-cp312-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "028a2624a734b28ebab84b1e6b3ae1306a74480d5b73182c0f7d78785530ba03",
"md5": "156a7d101532423f3f852e557ad08baa",
"sha256": "1a000cdb9b64b2241a9f9ea7fc1286781947b22a5727c3827d55383b7ef61073"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"has_sig": false,
"md5_digest": "156a7d101532423f3f852e557ad08baa",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 279838,
"upload_time": "2024-07-18T02:07:56",
"upload_time_iso_8601": "2024-07-18T02:07:56.659565Z",
"url": "https://files.pythonhosted.org/packages/02/8a/2624a734b28ebab84b1e6b3ae1306a74480d5b73182c0f7d78785530ba03/rebop-0.8.3-cp312-cp312-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "c9a23e6ba6dafc55e90482f844f82c5fb829d6358b72feae5f4131da8d93d0f3",
"md5": "980d6b117c5d5a7a364be1b001c494f0",
"sha256": "1bca10c482fd9671de454904ee26bcfdebcbdd50c4cfe547489471d70e3342df"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"has_sig": false,
"md5_digest": "980d6b117c5d5a7a364be1b001c494f0",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 293758,
"upload_time": "2024-07-18T02:08:02",
"upload_time_iso_8601": "2024-07-18T02:08:02.790752Z",
"url": "https://files.pythonhosted.org/packages/c9/a2/3e6ba6dafc55e90482f844f82c5fb829d6358b72feae5f4131da8d93d0f3/rebop-0.8.3-cp312-cp312-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "7411b37eea221dd7c5459541384c66c8d180b994a8dc05bf9416ce9b2186edb9",
"md5": "9557213074632df285e809b850a856f9",
"sha256": "18fb3650bb2ef0b4417d700da24e92222a48c3d5106f062aa845035c87b7ced4"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"has_sig": false,
"md5_digest": "9557213074632df285e809b850a856f9",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 355230,
"upload_time": "2024-07-18T02:08:08",
"upload_time_iso_8601": "2024-07-18T02:08:08.615215Z",
"url": "https://files.pythonhosted.org/packages/74/11/b37eea221dd7c5459541384c66c8d180b994a8dc05bf9416ce9b2186edb9/rebop-0.8.3-cp312-cp312-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "61fd3f4826e44d9af894ace88a9e0f0fc7e9047d8d224dce9174a5b722b41c7e",
"md5": "7d17202981e786f2a5c2faed75aa851b",
"sha256": "39b36be1f467cb1fad031a813d58ef6b0ae8beaf6de56001d5c882f52a07eec5"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"has_sig": false,
"md5_digest": "7d17202981e786f2a5c2faed75aa851b",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 266907,
"upload_time": "2024-07-18T02:08:19",
"upload_time_iso_8601": "2024-07-18T02:08:19.295442Z",
"url": "https://files.pythonhosted.org/packages/61/fd/3f4826e44d9af894ace88a9e0f0fc7e9047d8d224dce9174a5b722b41c7e/rebop-0.8.3-cp312-cp312-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "c81d9722c39f55f15ebca906671f7e1972260cde58fceda2c993532a7403c8b3",
"md5": "83ad6158afa13046fdc765b1ec2ea97e",
"sha256": "898e3271ab219e5eca37a1a04a92f8c992b931566d5e39c2a33ff701fcbd095c"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.whl",
"has_sig": false,
"md5_digest": "83ad6158afa13046fdc765b1ec2ea97e",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 275351,
"upload_time": "2024-07-18T02:08:13",
"upload_time_iso_8601": "2024-07-18T02:08:13.990525Z",
"url": "https://files.pythonhosted.org/packages/c8/1d/9722c39f55f15ebca906671f7e1972260cde58fceda2c993532a7403c8b3/rebop-0.8.3-cp312-cp312-manylinux_2_5_i686.manylinux1_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "1349a7304eb2c615317080ba882063af39a17b222ddb1635ffa7c965150f4a8f",
"md5": "83265a0c9388dc9479820f319a4ccfac",
"sha256": "48616e62d614aa0f57bd66b48dc5d2feae29ed1927527c6b523928572ab1ab53"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-musllinux_1_2_aarch64.whl",
"has_sig": false,
"md5_digest": "83265a0c9388dc9479820f319a4ccfac",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 451688,
"upload_time": "2024-07-18T02:08:32",
"upload_time_iso_8601": "2024-07-18T02:08:32.102127Z",
"url": "https://files.pythonhosted.org/packages/13/49/a7304eb2c615317080ba882063af39a17b222ddb1635ffa7c965150f4a8f/rebop-0.8.3-cp312-cp312-musllinux_1_2_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "060acb396d02b827be67b218093c0266b05d58983b9bc55980113c8aed2cbeb0",
"md5": "22e2ad7068458a91188f3a1c473d1b63",
"sha256": "19f612f628f139786645389d0baf2b8560c8ff7aef7e9e96fd33bf7eea0b1d86"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-musllinux_1_2_armv7l.whl",
"has_sig": false,
"md5_digest": "22e2ad7068458a91188f3a1c473d1b63",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 541478,
"upload_time": "2024-07-18T02:08:37",
"upload_time_iso_8601": "2024-07-18T02:08:37.933228Z",
"url": "https://files.pythonhosted.org/packages/06/0a/cb396d02b827be67b218093c0266b05d58983b9bc55980113c8aed2cbeb0/rebop-0.8.3-cp312-cp312-musllinux_1_2_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "98e3ea0767bfb61288adbd531e5778cf320b17855b0ad81022a998f0615c4b19",
"md5": "6ed22dda06753c8eaf0ff63c27ed80d8",
"sha256": "e65e03f4dea166825943c1aaa3758716c32363afafc0f842792d10da2e89f74a"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-musllinux_1_2_i686.whl",
"has_sig": false,
"md5_digest": "6ed22dda06753c8eaf0ff63c27ed80d8",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 455710,
"upload_time": "2024-07-18T02:08:43",
"upload_time_iso_8601": "2024-07-18T02:08:43.731174Z",
"url": "https://files.pythonhosted.org/packages/98/e3/ea0767bfb61288adbd531e5778cf320b17855b0ad81022a998f0615c4b19/rebop-0.8.3-cp312-cp312-musllinux_1_2_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "fa2f47ae9de1f8395890f33ad5e0c90bc5818a61b1e70e21917afb9bc52d8573",
"md5": "b6b3a95e0d31a69d569c0dc04a777d1a",
"sha256": "1ed93f712958ebb61aaf19141da2af25b15655317cde1843c5291bf199ec9c5d"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-cp312-musllinux_1_2_x86_64.whl",
"has_sig": false,
"md5_digest": "b6b3a95e0d31a69d569c0dc04a777d1a",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 437553,
"upload_time": "2024-07-18T02:08:49",
"upload_time_iso_8601": "2024-07-18T02:08:49.464402Z",
"url": "https://files.pythonhosted.org/packages/fa/2f/47ae9de1f8395890f33ad5e0c90bc5818a61b1e70e21917afb9bc52d8573/rebop-0.8.3-cp312-cp312-musllinux_1_2_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "56be25621c3dc814013fc62d934af039dc8a9d1bee2b68f190af9156959165f9",
"md5": "511c2e3bd11bbf58ced0dfa0b87d7baa",
"sha256": "b41b873c2a15bf8db508f21e17693380afe479568cb8866a3ec4498f97cf0fa0"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-none-win32.whl",
"has_sig": false,
"md5_digest": "511c2e3bd11bbf58ced0dfa0b87d7baa",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 147812,
"upload_time": "2024-07-18T02:08:58",
"upload_time_iso_8601": "2024-07-18T02:08:58.962706Z",
"url": "https://files.pythonhosted.org/packages/56/be/25621c3dc814013fc62d934af039dc8a9d1bee2b68f190af9156959165f9/rebop-0.8.3-cp312-none-win32.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "1d2048b5b18322718b9b8dfc5a773ddcc3578d3488e7436446c97beebc2f5fda",
"md5": "06baf4618f4e9268a4708a4562a7f73a",
"sha256": "ae463f9712c54ae9bda8e2988231c93d35bf24d2a96099b36bb6eb91a6a239ff"
},
"downloads": -1,
"filename": "rebop-0.8.3-cp312-none-win_amd64.whl",
"has_sig": false,
"md5_digest": "06baf4618f4e9268a4708a4562a7f73a",
"packagetype": "bdist_wheel",
"python_version": "cp312",
"requires_python": ">=3.10",
"size": 153535,
"upload_time": "2024-07-18T02:08:55",
"upload_time_iso_8601": "2024-07-18T02:08:55.212401Z",
"url": "https://files.pythonhosted.org/packages/1d/20/48b5b18322718b9b8dfc5a773ddcc3578d3488e7436446c97beebc2f5fda/rebop-0.8.3-cp312-none-win_amd64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "1d8f8e0dbcd22ef1a9bae8f929185ae78eba129602b6548f0ea9cc117a1c5ecf",
"md5": "72fcb3e27a2907ea3281bd23c9e7cb30",
"sha256": "4aa01a852821e24f882ed33367f853b3b64e87feadab69bb239cbcf4f473812d"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"has_sig": false,
"md5_digest": "72fcb3e27a2907ea3281bd23c9e7cb30",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 273227,
"upload_time": "2024-07-18T02:07:51",
"upload_time_iso_8601": "2024-07-18T02:07:51.638988Z",
"url": "https://files.pythonhosted.org/packages/1d/8f/8e0dbcd22ef1a9bae8f929185ae78eba129602b6548f0ea9cc117a1c5ecf/rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_aarch64.manylinux2014_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "58bd1404432044a2423d2d866e0959aba051dee1922af1d2d6005eff2c215003",
"md5": "9e1ce8102dd906f4c3531a28fafe5ba3",
"sha256": "bba2d935633cb950828fffcb5cf38a5a45ba090bc530c69f78109e6b6965e4b4"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"has_sig": false,
"md5_digest": "9e1ce8102dd906f4c3531a28fafe5ba3",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 280535,
"upload_time": "2024-07-18T02:07:58",
"upload_time_iso_8601": "2024-07-18T02:07:58.195260Z",
"url": "https://files.pythonhosted.org/packages/58/bd/1404432044a2423d2d866e0959aba051dee1922af1d2d6005eff2c215003/rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_armv7l.manylinux2014_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "74fbd42515155a907ae85094ac4cdcb325b55db5146a68eb509d115a88f11f76",
"md5": "bc1ac4198f6da303e2fa6b31be2b3027",
"sha256": "418cff3b8d4e44cc3b86002566e162549c001ded7a5756ccb188f8ec0b81d1d2"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"has_sig": false,
"md5_digest": "bc1ac4198f6da303e2fa6b31be2b3027",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 294944,
"upload_time": "2024-07-18T02:08:04",
"upload_time_iso_8601": "2024-07-18T02:08:04.419503Z",
"url": "https://files.pythonhosted.org/packages/74/fb/d42515155a907ae85094ac4cdcb325b55db5146a68eb509d115a88f11f76/rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_ppc64le.manylinux2014_ppc64le.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "6030b245a43e28ea9542f4dc57ba0f14e2fd93661826eff309628b87dd77020e",
"md5": "bad6c4de8c68113c99b18a5e50efc98b",
"sha256": "8561c384c4261b9539300e9f19b268c3218da5a2092fff48a9f40e3dfc667631"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"has_sig": false,
"md5_digest": "bad6c4de8c68113c99b18a5e50efc98b",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 352399,
"upload_time": "2024-07-18T02:08:10",
"upload_time_iso_8601": "2024-07-18T02:08:10.183277Z",
"url": "https://files.pythonhosted.org/packages/60/30/b245a43e28ea9542f4dc57ba0f14e2fd93661826eff309628b87dd77020e/rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_s390x.manylinux2014_s390x.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "a27b6bc95fe500b7bd05bf428454cf9e8d4dcf1954dfccb1a1d3a5ee49fe1f0c",
"md5": "7eabefed09ee585847c7a6ad49f5381f",
"sha256": "363d888996d0d448c5d2d4cadd5e6650e7b433fd4f317334727a3f237741801f"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"has_sig": false,
"md5_digest": "7eabefed09ee585847c7a6ad49f5381f",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 267549,
"upload_time": "2024-07-18T02:08:20",
"upload_time_iso_8601": "2024-07-18T02:08:20.516375Z",
"url": "https://files.pythonhosted.org/packages/a2/7b/6bc95fe500b7bd05bf428454cf9e8d4dcf1954dfccb1a1d3a5ee49fe1f0c/rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_17_x86_64.manylinux2014_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "40ed6504c25b85e45b3077b0ae8ff3a508dc48527f906591a45a5ca8d5a6b3b4",
"md5": "0475daa3544842470db162a5b6bddad8",
"sha256": "28ab57af5fd9c91bccaf8e2731df3bfd45936c84aca2898aebb78141112d4816"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_5_i686.manylinux1_i686.whl",
"has_sig": false,
"md5_digest": "0475daa3544842470db162a5b6bddad8",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 276339,
"upload_time": "2024-07-18T02:08:15",
"upload_time_iso_8601": "2024-07-18T02:08:15.280599Z",
"url": "https://files.pythonhosted.org/packages/40/ed/6504c25b85e45b3077b0ae8ff3a508dc48527f906591a45a5ca8d5a6b3b4/rebop-0.8.3-pp310-pypy310_pp73-manylinux_2_5_i686.manylinux1_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "be2348bfc03b9ae195075576a5c9ec0ee9c4103fce0f34dbe8dc145097bafe29",
"md5": "952824a99f3ab8c958f9adf317112893",
"sha256": "fe8c5967027edadc7e8aa1b064a01340f9faedf3bf948df315200816f859aaa4"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_aarch64.whl",
"has_sig": false,
"md5_digest": "952824a99f3ab8c958f9adf317112893",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 452587,
"upload_time": "2024-07-18T02:08:33",
"upload_time_iso_8601": "2024-07-18T02:08:33.431147Z",
"url": "https://files.pythonhosted.org/packages/be/23/48bfc03b9ae195075576a5c9ec0ee9c4103fce0f34dbe8dc145097bafe29/rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_aarch64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "c4e9b6c92ab296f9db0f629d32230f9170d1902a57fe1409e4f9b1fac8464c62",
"md5": "1dcfcbadd62849532aef79c61e6f7c43",
"sha256": "dd2090a556f7a01932857b3c39e102d5008cabcb5db596e60938ed22b40b613c"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_armv7l.whl",
"has_sig": false,
"md5_digest": "1dcfcbadd62849532aef79c61e6f7c43",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 542467,
"upload_time": "2024-07-18T02:08:39",
"upload_time_iso_8601": "2024-07-18T02:08:39.631824Z",
"url": "https://files.pythonhosted.org/packages/c4/e9/b6c92ab296f9db0f629d32230f9170d1902a57fe1409e4f9b1fac8464c62/rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_armv7l.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "e677d86ca87f5a4d1944dea440e191ed78b52801e0db23617bd7a99b84916d19",
"md5": "1bf6e19ff1e8bf635e73420e83f959ae",
"sha256": "fea828a0550056276a00861e3911df7c1eda21fe6f294845c029fa3a2294c9d0"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_i686.whl",
"has_sig": false,
"md5_digest": "1bf6e19ff1e8bf635e73420e83f959ae",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 456321,
"upload_time": "2024-07-18T02:08:44",
"upload_time_iso_8601": "2024-07-18T02:08:44.970521Z",
"url": "https://files.pythonhosted.org/packages/e6/77/d86ca87f5a4d1944dea440e191ed78b52801e0db23617bd7a99b84916d19/rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_i686.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "0ed3fe60d44cb975ba28439d7545bee04bff8db6cfac02d49590e05bf5543547",
"md5": "56e061a5cbf3126686bf9f265f62f4df",
"sha256": "02b1f4ed6bb309bc12c12e548e35c4bf9c2db945144c56b2cfd558bc4ebfade4"
},
"downloads": -1,
"filename": "rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_x86_64.whl",
"has_sig": false,
"md5_digest": "56e061a5cbf3126686bf9f265f62f4df",
"packagetype": "bdist_wheel",
"python_version": "pp310",
"requires_python": ">=3.10",
"size": 438238,
"upload_time": "2024-07-18T02:08:50",
"upload_time_iso_8601": "2024-07-18T02:08:50.717637Z",
"url": "https://files.pythonhosted.org/packages/0e/d3/fe60d44cb975ba28439d7545bee04bff8db6cfac02d49590e05bf5543547/rebop-0.8.3-pp310-pypy310_pp73-musllinux_1_2_x86_64.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "3e2a02c523a894dc9d5a20df4bc225610b56e0f8e3ab717b03f2421d379d1a75",
"md5": "56e6c7a9c3d5163002ddbe0dec171f16",
"sha256": "ffe4df61ccef9b4d829dcbaffd1e137e07909c88068b3e7373ccd1c68a6af768"
},
"downloads": -1,
"filename": "rebop-0.8.3.tar.gz",
"has_sig": false,
"md5_digest": "56e6c7a9c3d5163002ddbe0dec171f16",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 115941,
"upload_time": "2024-07-18T02:08:52",
"upload_time_iso_8601": "2024-07-18T02:08:52.057433Z",
"url": "https://files.pythonhosted.org/packages/3e/2a/02c523a894dc9d5a20df4bc225610b56e0f8e3ab717b03f2421d379d1a75/rebop-0.8.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-07-18 02:08:52",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "Armavica",
"github_project": "rebop",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "rebop"
}
