<p align="center">
<img width="700" height="300" src="https://raw.githubusercontent.com/flowersteam/sbmltoodejax/main/docs/source/_static/logo.svg" alt="SBMLtoODEjax's logo">
</p>
[](https://badge.fury.io/py/SBMLtoODEjax)
[](https://pepy.tech/project/sbmltoodejax)
[](https://arxiv.org/abs/2307.08452)
# About
SBMLtoODEjax is a lightweight library that allows to automatically parse and convert SBML models into python models
written end-to-end in [JAX](https://github.com/google/jax),
a high-performance numerical computing library with automatic differentiation capabilities.
SBMLtoODEjax is targeted at researchers that aim to incorporate SBML-specified ordinary differential equation (ODE)
models into their python projects and machine learning pipelines,
in order to perform efficient numerical simulation and optimization with only a few lines of code (by taking advantage
of JAX’s core transformation features).
SBMLtoODEjax extends [SBMLtoODEpy](https://github.com/AnabelSMRuggiero/sbmltoodepy), a python library developed in 2019
for converting SBML files into python files written in Numpy/Scipy.
The chosen conventions for the generated variables and modules are slightly different from the standard SBML
conventions (used in the SBMLtoODEpy library)
with the aim here to accommodate for more flexible manipulations while preserving JAX-like functional programming style.
> *👉 In short, SBMLtoODEjax facilitates the re-use of biological network models and their manipulation in python projects
while tailoring them to take advantage of JAX main features for efficient and parallel computations.*
> 📖 The documentation, notebook tutorials and public APU are available at https://developmentalsystems.org/sbmltoodejax/.
# Installation
The latest stable release of `SBMLtoODEjax` can be installed via `pip`:
```bash
pip install sbmltoodejax
```
Requires SBMLtoODEpy, JAX (cpu) and Equinox.
# Why use SBMLtoODEjax?
## Simplicity and extensibility
SBMLtoODEjax retains the simplicity of the original [SBMLtoODEPy](https://github.com/AnabelSMRuggiero/sbmltoodepy) library to facilitate incorporation and refactoring of the
ODE models into one’s own python
projects. As shown below, with only a few lines of python code one can load and simulate existing SBML
files.
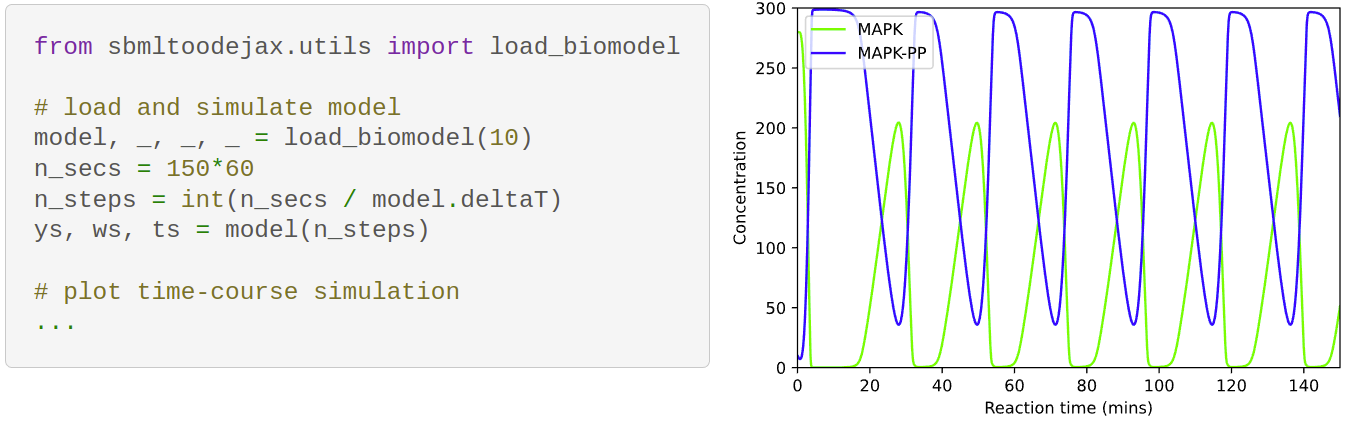
<font size="2" color="gray" > Example code (left) and output snapshot (right) reproducing [original simulation results](https://www.ebi.ac.uk/biomodels/BIOMD0000000010#Curation)
of Kholodenko 2000's paper hosted on BioModels website.
</font>
> **👉 Check our [Numerical Simulation](tutorials/biomodels_curation.ipynb) tutorial to reproduce results yourself and see more examples.**
## JAX-friendly
The generated python models are tailored to take advantage of JAX main features.
```python
class ModelRollout(eqx.Module):
def __call__(self, n_steps, y0, w0, c, t0=0.0):
@jit # use of jit transformation decorator
def f(carry, x):
y, w, c, t = carry
return self.modelstepfunc(y, w, c, t, self.deltaT), (y, w, t)
# use of scan primitive to replace for loop (and reduce compilation time)
(y, w, c, t), (ys, ws, ts) = lax.scan(f, (y0, w0, c, t0), jnp.arange(n_steps))
ys = jnp.moveaxis(ys, 0, -1)
ws = jnp.moveaxis(ws, 0, -1)
return ys, ws, ts
```
As shown above, model rollouts use `jit` transformation and `scan` primitive to reduce compilation and execution time of
the recursive ODE integration steps, which is particularly useful when running large numbers of steps (long reaction
times). Models also inherit from the [Equinox module abstraction](https://docs.kidger.site/equinox/api/module/module/) and are registered as PyTree
containers, which facilitates the application of JAX core transformations to any SBMLtoODEjax object.
## Efficiency simulation and optimization
The application of JAX core transformations, such as just-in-time
compilation (`jit`), automatic vectorization (`vmap`) and automatic differentiation (`grad`), to the generated models make it very easy (and
seamless) to efficiently run simulations in parallel.
For instance, as shown below, with only a few lines of python
code one can vectorize calls to model rollout and perform batched computations efficiently, which is particularly useful when
considering large batch sizes.
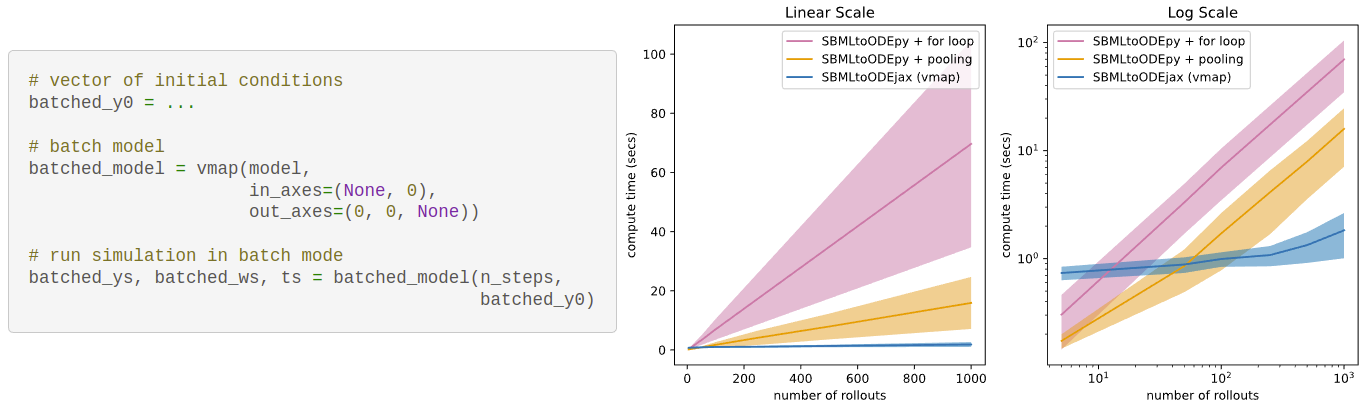
<font size="2" color="gray"> (left) Example code to vectorize calls to model rollout
(right) Results of a (rudimentary) benchmark comparing the average simulation time of models implemented with SBMLtoODEpy
versus SBMLtoODEjax (for different number of rollouts i.e. batch size).
</font>
> **👉 Check our [Benchmarking](https://developmentalsystems.org/sbmltoodejax/tutorials/benchmark.html) notebook for additional details on the benchmark results**.
Finally, as shown below, SBMLtoODEjax models can also be integrated within [Optax](https://github.com/deepmind/optax) pipelines,
a gradient processing and optimization library for [JAX](https://github.com/google/jax), allowing to optimize model parameters and/or
external interventions with stochastic gradient descent.
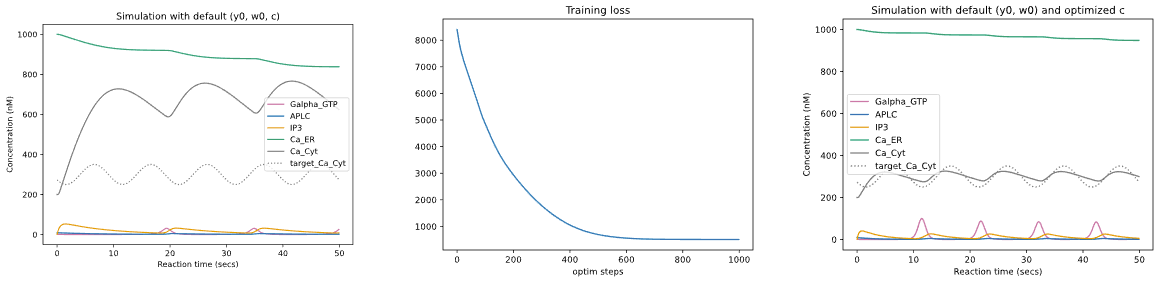
<font size="2" color="gray"> (left) Default simulation results of [biomodel #145](https://www.ebi.ac.uk/biomodels/BIOMD0000000145) which models ATP-induced intracellular calcium oscillations,
and target sine-wave pattern for Ca_Cyt concentration.
(middle) Training loss obtained when running the Optax optimization loop, with Adam optimizer, over the model kinematic parameters *c*.
(right) Simulation results obtained after optimization.
</font>
> **👉 Check our [Gradient Descent](https://developmentalsystems.org/sbmltoodejax/tutorials/gradient_descent.html) tutorial to reproduce the result yourself and try more-advanced optimization usages.**
# All contributions are welcome!
SBMLtoODEjax is in its early stage and any sort of contribution will be highly appreciated.
## Suggested contributions
They are several use cases that are not handled by the current codebase including:
1) **Events**: SBML files with events (discrete occurrences that can trigger discontinuous changes in the model) are not handled
2) **Math Functions**: we handle a large portion, but not all, of functions possibly-used in SBML files (see `mathFuncs` in `sbmltoodejax.modulegeneration.GenerateModel`)
3) **Custom solvers**: To integrate the model's equation, we use jax experimental `odeint` solver but do not yet allow for other solvers.
4) **NaN/Negative values**: numerical simulation sometimes leads to NaN values (or negative values for the species amounts) which could either be due to wrong parsing or solver issues
This means that a large portion of the possible SBML files cannot yet be simulated, for instance as we detail on the below image, out of 1048
curated models that one can load from the BioModels website, only 232 can successfully be simulated (given the default initial conditions) in SBMLtoODEjax:
<img src="https://raw.githubusercontent.com/flowersteam/sbmltoodejax/main/docs/source/_static/error_cases.png" width="450px">
> 👉 Please consider contributing and check our [Contribution Guidelines](https://developmentalsystems.org/sbmltoodejax/contributing.html) to learn how to do so.
# License
The SBMLtoODEjax project is licensed under
the [MIT license](https://github.com/flowersteam/sbmltoodejax/blob/main/LICENSE).
# Acknowledgements
SBMLtoODEjax builds on:
* [SBMLtoODEpy](https://github.com/AnabelSMRuggiero/sbmltoodepy)'s parsing and conversion of SBML files, by Steve M.
Ruggiero and Ashlee N. Ford
* [JAX](https://github.com/google/jax)'s composable transformations, by the Google team
* [Equinox](https://github.com/patrick-kidger/equinox)'s module abstraction, by Patrick Kidger
* [BasiCO](https://github.com/copasi/basico/blob/d058c10dd51f2c3e926efeaa29c6194f86bfdc90/basico/biomodels.py)'s access
the BioModels REST api, by the COPASI team
Our documentation was also inspired by the [GPJax](https://docs.jaxgaussianprocesses.com/) documentation, by Thomas
Pinder and team.
# Citing SBMLtoODEjax
If you use SBMLtoODEjax in your research, please cite the paper:
```
@inproceedings{etcheverry2023sbmltoodejax,
title={SBMLtoODEjax: Efficient Simulation and Optimization of Biological Network Models in JAX},
author={Mayalen Etcheverry and Michael Levin and Clement Moulin-Frier and Pierre-Yves Oudeyer},
booktitle={NeurIPS 2023 AI for Science Workshop},
year={2023},
url={https://openreview.net/forum?id=exP6UntwqJ}
}
```
Raw data
{
"_id": null,
"home_page": "https://github.com/flowersteam/sbmltoodejax",
"name": "sbmltoodejax",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "",
"author": "Mayalen Etcheverry",
"author_email": "mayalen.etcheverry@inria.fr",
"download_url": "https://files.pythonhosted.org/packages/4b/c8/8eeb90011592916fc10211e49ead71193465e6afcfd2cadcbb9dd12ab7ff/sbmltoodejax-0.4.2.tar.gz",
"platform": null,
"description": "<p align=\"center\">\n<img width=\"700\" height=\"300\" src=\"https://raw.githubusercontent.com/flowersteam/sbmltoodejax/main/docs/source/_static/logo.svg\" alt=\"SBMLtoODEjax's logo\">\n</p>\n\n[](https://badge.fury.io/py/SBMLtoODEjax)\n[](https://pepy.tech/project/sbmltoodejax)\n[](https://arxiv.org/abs/2307.08452)\n\n# About\n\nSBMLtoODEjax is a lightweight library that allows to automatically parse and convert SBML models into python models\nwritten end-to-end in [JAX](https://github.com/google/jax),\na high-performance numerical computing library with automatic differentiation capabilities.\nSBMLtoODEjax is targeted at researchers that aim to incorporate SBML-specified ordinary differential equation (ODE)\nmodels into their python projects and machine learning pipelines,\nin order to perform efficient numerical simulation and optimization with only a few lines of code (by taking advantage\nof JAX\u2019s core transformation features).\n\nSBMLtoODEjax extends [SBMLtoODEpy](https://github.com/AnabelSMRuggiero/sbmltoodepy), a python library developed in 2019\nfor converting SBML files into python files written in Numpy/Scipy.\nThe chosen conventions for the generated variables and modules are slightly different from the standard SBML\nconventions (used in the SBMLtoODEpy library)\nwith the aim here to accommodate for more flexible manipulations while preserving JAX-like functional programming style.\n\n> *\ud83d\udc49 In short, SBMLtoODEjax facilitates the re-use of biological network models and their manipulation in python projects\nwhile tailoring them to take advantage of JAX main features for efficient and parallel computations.*\n\n> \ud83d\udcd6 The documentation, notebook tutorials and public APU are available at https://developmentalsystems.org/sbmltoodejax/.\n\n# Installation\n\nThe latest stable release of `SBMLtoODEjax` can be installed via `pip`:\n\n```bash\npip install sbmltoodejax\n```\n\nRequires SBMLtoODEpy, JAX (cpu) and Equinox.\n\n# Why use SBMLtoODEjax?\n\n## Simplicity and extensibility\nSBMLtoODEjax retains the simplicity of the original [SBMLtoODEPy](https://github.com/AnabelSMRuggiero/sbmltoodepy) library to facilitate incorporation and refactoring of the\nODE models into one\u2019s own python\nprojects. As shown below, with only a few lines of python code one can load and simulate existing SBML\nfiles.\n\n\n<font size=\"2\" color=\"gray\" > Example code (left) and output snapshot (right) reproducing [original simulation results](https://www.ebi.ac.uk/biomodels/BIOMD0000000010#Curation)\nof Kholodenko 2000's paper hosted on BioModels website.\n</font>\n\n> **\ud83d\udc49 Check our [Numerical Simulation](tutorials/biomodels_curation.ipynb) tutorial to reproduce results yourself and see more examples.**\n\n\n## JAX-friendly\nThe generated python models are tailored to take advantage of JAX main features.\n```python\nclass ModelRollout(eqx.Module):\n \n def __call__(self, n_steps, y0, w0, c, t0=0.0):\n\n @jit # use of jit transformation decorator\n def f(carry, x):\n y, w, c, t = carry\n return self.modelstepfunc(y, w, c, t, self.deltaT), (y, w, t)\n \n # use of scan primitive to replace for loop (and reduce compilation time)\n (y, w, c, t), (ys, ws, ts) = lax.scan(f, (y0, w0, c, t0), jnp.arange(n_steps)) \n ys = jnp.moveaxis(ys, 0, -1)\n ws = jnp.moveaxis(ws, 0, -1)\n \n return ys, ws, ts\n```\nAs shown above, model rollouts use `jit` transformation and `scan` primitive to reduce compilation and execution time of\nthe recursive ODE integration steps, which is particularly useful when running large numbers of steps (long reaction\ntimes). Models also inherit from the [Equinox module abstraction](https://docs.kidger.site/equinox/api/module/module/) and are registered as PyTree\ncontainers, which facilitates the application of JAX core transformations to any SBMLtoODEjax object.\n\n## Efficiency simulation and optimization\nThe application of JAX core transformations, such as just-in-time\ncompilation (`jit`), automatic vectorization (`vmap`) and automatic differentiation (`grad`), to the generated models make it very easy (and\nseamless) to efficiently run simulations in parallel. \n\nFor instance, as shown below, with only a few lines of python\ncode one can vectorize calls to model rollout and perform batched computations efficiently, which is particularly useful when\nconsidering large batch sizes.\n\n <font size=\"2\" color=\"gray\"> (left) Example code to vectorize calls to model rollout\n(right) Results of a (rudimentary) benchmark comparing the average simulation time of models implemented with SBMLtoODEpy\nversus SBMLtoODEjax (for different number of rollouts i.e. batch size).\n</font>\n\n> **\ud83d\udc49 Check our [Benchmarking](https://developmentalsystems.org/sbmltoodejax/tutorials/benchmark.html) notebook for additional details on the benchmark results**.\n\n\nFinally, as shown below, SBMLtoODEjax models can also be integrated within [Optax](https://github.com/deepmind/optax) pipelines,\na gradient processing and optimization library for [JAX](https://github.com/google/jax), allowing to optimize model parameters and/or\nexternal interventions with stochastic gradient descent.\n\n\n<font size=\"2\" color=\"gray\"> (left) Default simulation results of [biomodel #145](https://www.ebi.ac.uk/biomodels/BIOMD0000000145) which models ATP-induced intracellular calcium oscillations,\nand target sine-wave pattern for Ca_Cyt concentration.\n(middle) Training loss obtained when running the Optax optimization loop, with Adam optimizer, over the model kinematic parameters *c*.\n(right) Simulation results obtained after optimization. \n</font>\n\n> **\ud83d\udc49 Check our [Gradient Descent](https://developmentalsystems.org/sbmltoodejax/tutorials/gradient_descent.html) tutorial to reproduce the result yourself and try more-advanced optimization usages.**\n\n# All contributions are welcome!\n\nSBMLtoODEjax is in its early stage and any sort of contribution will be highly appreciated.\n\n\n## Suggested contributions\nThey are several use cases that are not handled by the current codebase including:\n1) **Events**: SBML files with events (discrete occurrences that can trigger discontinuous changes in the model) are not handled\n2) **Math Functions**: we handle a large portion, but not all, of functions possibly-used in SBML files (see `mathFuncs` in `sbmltoodejax.modulegeneration.GenerateModel`)\n3) **Custom solvers**: To integrate the model's equation, we use jax experimental `odeint` solver but do not yet allow for other solvers.\n4) **NaN/Negative values**: numerical simulation sometimes leads to NaN values (or negative values for the species amounts) which could either be due to wrong parsing or solver issues\n\nThis means that a large portion of the possible SBML files cannot yet be simulated, for instance as we detail on the below image, out of 1048\ncurated models that one can load from the BioModels website, only 232 can successfully be simulated (given the default initial conditions) in SBMLtoODEjax:\n<img src=\"https://raw.githubusercontent.com/flowersteam/sbmltoodejax/main/docs/source/_static/error_cases.png\" width=\"450px\">\n\n> \ud83d\udc49 Please consider contributing and check our [Contribution Guidelines](https://developmentalsystems.org/sbmltoodejax/contributing.html) to learn how to do so.\n\n# License\n\nThe SBMLtoODEjax project is licensed under\nthe [MIT license](https://github.com/flowersteam/sbmltoodejax/blob/main/LICENSE).\n\n# Acknowledgements\n\nSBMLtoODEjax builds on:\n\n* [SBMLtoODEpy](https://github.com/AnabelSMRuggiero/sbmltoodepy)'s parsing and conversion of SBML files, by Steve M.\n Ruggiero and Ashlee N. Ford\n* [JAX](https://github.com/google/jax)'s composable transformations, by the Google team\n* [Equinox](https://github.com/patrick-kidger/equinox)'s module abstraction, by Patrick Kidger\n* [BasiCO](https://github.com/copasi/basico/blob/d058c10dd51f2c3e926efeaa29c6194f86bfdc90/basico/biomodels.py)'s access\n the BioModels REST api, by the COPASI team\n\nOur documentation was also inspired by the [GPJax](https://docs.jaxgaussianprocesses.com/) documentation, by Thomas\nPinder and team.\n\n# Citing SBMLtoODEjax\n\nIf you use SBMLtoODEjax in your research, please cite the paper:\n\n```\n@inproceedings{etcheverry2023sbmltoodejax,\ntitle={SBMLtoODEjax: Efficient Simulation and Optimization of Biological Network Models in JAX},\nauthor={Mayalen Etcheverry and Michael Levin and Clement Moulin-Frier and Pierre-Yves Oudeyer},\nbooktitle={NeurIPS 2023 AI for Science Workshop},\nyear={2023},\nurl={https://openreview.net/forum?id=exP6UntwqJ}\n}\n```\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "lightweight library that allows to automatically parse and convert SBML models into python models written end-to-end in JAX",
"version": "0.4.2",
"project_urls": {
"Homepage": "https://github.com/flowersteam/sbmltoodejax"
},
"split_keywords": [],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "2691c0787c41f2bd4d8dba132d112d2a60af1bcae19687c9cbd17339a6f130f6",
"md5": "7fcb12e76629feb2c4bf5f2c9d538903",
"sha256": "c055fa0fa8ce1d833e20a12b0cbc8df2d5b99639546b99997cb1efbe1500cd22"
},
"downloads": -1,
"filename": "sbmltoodejax-0.4.2-py3-none-any.whl",
"has_sig": false,
"md5_digest": "7fcb12e76629feb2c4bf5f2c9d538903",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": null,
"size": 17589,
"upload_time": "2024-02-26T16:27:32",
"upload_time_iso_8601": "2024-02-26T16:27:32.908786Z",
"url": "https://files.pythonhosted.org/packages/26/91/c0787c41f2bd4d8dba132d112d2a60af1bcae19687c9cbd17339a6f130f6/sbmltoodejax-0.4.2-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "4bc88eeb90011592916fc10211e49ead71193465e6afcfd2cadcbb9dd12ab7ff",
"md5": "8774d93a6ef7a56e42867c532a0d9237",
"sha256": "af982fdae54808231d4d238b60da6fc5173fe395b3dc373ee0bac7c0d0100685"
},
"downloads": -1,
"filename": "sbmltoodejax-0.4.2.tar.gz",
"has_sig": false,
"md5_digest": "8774d93a6ef7a56e42867c532a0d9237",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 20267,
"upload_time": "2024-02-26T16:27:34",
"upload_time_iso_8601": "2024-02-26T16:27:34.576768Z",
"url": "https://files.pythonhosted.org/packages/4b/c8/8eeb90011592916fc10211e49ead71193465e6afcfd2cadcbb9dd12ab7ff/sbmltoodejax-0.4.2.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-02-26 16:27:34",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "flowersteam",
"github_project": "sbmltoodejax",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "sbmltoodejax"
}
