# Star Alleles Comparator (star_allele_comp)
The comparator can compare HLA or KIR alleles between cohorts
## Install
``` bash
pip install git+https://github.com/linnil1/star_alleles_comparator
```
## Usage
### 1. Using command line
``` bash
star_allele_comp hla_result1.csv hla_result2.csv --family hla --save tmp --plot -v
```
The results will output to screen and save in .txt and .csv format.
The example output is the same as below (see next section).
The input CSV should adhere to the following format:
Columns
* `id` (**required**): The sample ID.
* `method` (**optinal**): The method. If not specified, filename will be used.
* `allele*` (**required**) Columns starting with `allele` will be used to store the allele for each id/sample with corresponding method.
The value can be NULL, empty.
#### Format 1: Separate Columns for Alleles
For this format, each allele is represented in separate columns:
``` csv
method,id,allele1,allele2,allele3,allele4
method1,id1,"A*01:02:03:04","A*01:02","B*01:01:01:01"
method1,id2,"A*01:02:03:04","A*01:02","B*01:01:01:01"
method1,id2,"C*03","C*03:03"
method2,id1,"A*01:02:03:04","A*01:02:03","B*01:02:02:01","B*04:01:02"
method2,id2,"A*01:02:03:04","A*01:02:03","B*01:02:02:01","B*04:01:02"
method2,id2,"C*03:03", "C*03:02"
```
#### Format 2: Using "alleles" Column with Underscore as Separator
In this format, the `alleles` column contains a single string with alleles separated by underscores:
``` csv
method,id,alleles
method1,id3,"KIR2DL1*0010203_KIR2DL1*001_KIR2DS1*0010101"
method1,id4,"KIR2DL1*0010203_KIR2DL1*00102_KIR2DS1*00101"
method1,id3,"KIR2DL1*03105_KIR2DL1*03:03"
method2,id3,"KIR2DL1*001_KIR2DL1*0030203_KIR2DS1*0010208_KIR2DS1*0040102"
method2,id4,"KIR2DL1*0010203_KIR2DL1*0010203_KIR2DS1*0010202_KIR2DS1*0040302"
method2,id4,"KIR2DL1*00303_KIR2DL1*03002"
```
### 2. Using Python functions
#### Run comparison
``` python
from star_allele_comp import compare_method, print_all_summary, plot_summary
cohort = {
"method1": { "sample_id1": [ "A*01:02:03:04", "A*01:02", "B*01:01:01:01", "B*03:01"] },
"method2": { "sample_id1": [ "A*01:02:03:04", "A*01:02:03", "B*01:02:02:01", "B*04:01:02"] },
}
ground_truth_method = "method1"
result = compare_method(cohort, ground_truth_method, "hla")
```
#### Print result allele by allele
``` python
print(result)
# Method method2
# Sample sample_id1
# A*01:02:03:04 =4= A*01:02:03:04
# A*01:02 =2= A*01:02:03
# B*01:01:01:01 =1= B*01:02:02:01
# B*03:01 =0= B*04:01:02
# Note:
# Left hand side is the alleles in reference method/cohort
# Right hand side is the allele in another method/cohort
```
#### Print summary (i.e. Accuracy vs Resolution, Confusion Matrix)
``` python
# details are in star_allele_comp/summary.py:print_all_summary
df_cohort = result.to_dataframe()
print_all_summary(df_cohort)
```
``` txt
Accuracy summary
Accuracy num_match num_ref
Resolution 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN
method
method1 1.0 1.00 1.0 1.0 1.0 0.0 0.0 4 4 4 2 2 0 0 4 4 4 2 2 0 0
method2 1.0 0.75 0.5 0.5 0.5 0.0 0.0 4 3 2 1 1 0 0 4 4 4 2 2 0 0
# Note In the accuracy summary table:
# * num_match represents the number of alleles that match the alleles in the ground truth method under the specific `Resolution`.
# * num_ref indicates the number of reference alleles with resolution >= `Resolution`
# * Accuracy is calculated as the ratio of num_match to num_ref.
# * Accuracy in FP is False Discovery Rate (FDR)
# * Accuracy in FN is False Negative Rate (FNR)
Confusion matrix (not the same sample)
Count
match_res -1 0 1 2 3
ref_res
-1 2 0 0 0 0
1 1 1 0 0 0
2 1 0 2 6 0
3 0 0 0 0 1
4 0 0 0 0 1
# Note
# -1 indicates FP or FN
Accuracy summary per resolution per gene
Accuracy num_match num_ref
Resolution 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN
method gene
method1 A 1.0 1.0 1.0 1.0 1.0 0.0 0.0 2 2 2 1 1 0 0 2 2 2 1 1 0 0
B 1.0 1.0 1.0 1.0 1.0 0.0 0.0 2 2 2 1 1 0 0 2 2 2 1 1 0 0
```
#### Plot summary (i.e. Accuracy vs Resolution, gene, methods)
``` python
figs = plot_summary(df_cohort)
# You can use Dash to show it
from dash import dcc, html, Dash
app = Dash(__name__)
app.layout = html.Div(children=[dcc.Graph(figure=fig) for fig in figs])
app.run(debug=True)
```
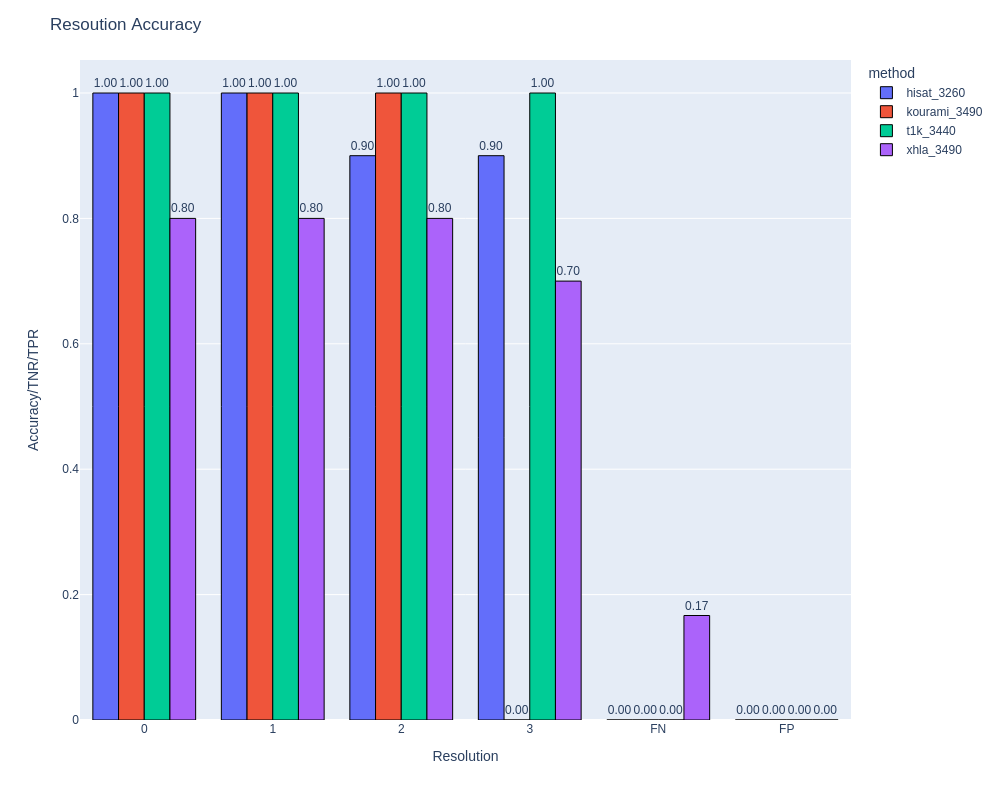
## Develop
``` bash
pip install pdoc
pdoc star_allele_comp --docformat google
```
## Details


Raw data
{
"_id": null,
"home_page": "",
"name": "star-allele-comp",
"maintainer": "",
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": "",
"keywords": "HLA,KIR,comparator,allele,star-allele",
"author": "linnil1",
"author_email": "",
"download_url": "https://files.pythonhosted.org/packages/99/c4/dd100bfb49e23877cdc89e4518e2baba69e290ce8d79928f59c3c090036c/star_allele_comp-0.2.tar.gz",
"platform": null,
"description": "# Star Alleles Comparator (star_allele_comp)\n\nThe comparator can compare HLA or KIR alleles between cohorts\n\n\n## Install\n\n``` bash\npip install git+https://github.com/linnil1/star_alleles_comparator\n```\n\n\n## Usage\n\n### 1. Using command line\n\n``` bash\nstar_allele_comp hla_result1.csv hla_result2.csv --family hla --save tmp --plot -v\n```\n\nThe results will output to screen and save in .txt and .csv format.\n\nThe example output is the same as below (see next section).\n\nThe input CSV should adhere to the following format:\n\nColumns\n* `id` (**required**): The sample ID.\n* `method` (**optinal**): The method. If not specified, filename will be used.\n* `allele*` (**required**) Columns starting with `allele` will be used to store the allele for each id/sample with corresponding method.\n The value can be NULL, empty.\n\n\n#### Format 1: Separate Columns for Alleles\n\nFor this format, each allele is represented in separate columns:\n\n``` csv\nmethod,id,allele1,allele2,allele3,allele4\nmethod1,id1,\"A*01:02:03:04\",\"A*01:02\",\"B*01:01:01:01\"\nmethod1,id2,\"A*01:02:03:04\",\"A*01:02\",\"B*01:01:01:01\"\nmethod1,id2,\"C*03\",\"C*03:03\"\nmethod2,id1,\"A*01:02:03:04\",\"A*01:02:03\",\"B*01:02:02:01\",\"B*04:01:02\"\nmethod2,id2,\"A*01:02:03:04\",\"A*01:02:03\",\"B*01:02:02:01\",\"B*04:01:02\"\nmethod2,id2,\"C*03:03\", \"C*03:02\"\n```\n\n#### Format 2: Using \"alleles\" Column with Underscore as Separator\n\nIn this format, the `alleles` column contains a single string with alleles separated by underscores:\n\n``` csv\nmethod,id,alleles\nmethod1,id3,\"KIR2DL1*0010203_KIR2DL1*001_KIR2DS1*0010101\"\nmethod1,id4,\"KIR2DL1*0010203_KIR2DL1*00102_KIR2DS1*00101\"\nmethod1,id3,\"KIR2DL1*03105_KIR2DL1*03:03\"\nmethod2,id3,\"KIR2DL1*001_KIR2DL1*0030203_KIR2DS1*0010208_KIR2DS1*0040102\"\nmethod2,id4,\"KIR2DL1*0010203_KIR2DL1*0010203_KIR2DS1*0010202_KIR2DS1*0040302\"\nmethod2,id4,\"KIR2DL1*00303_KIR2DL1*03002\"\n```\n\n\n### 2. Using Python functions\n\n#### Run comparison\n\n``` python\nfrom star_allele_comp import compare_method, print_all_summary, plot_summary\ncohort = {\n \"method1\": { \"sample_id1\": [ \"A*01:02:03:04\", \"A*01:02\", \"B*01:01:01:01\", \"B*03:01\"] },\n \"method2\": { \"sample_id1\": [ \"A*01:02:03:04\", \"A*01:02:03\", \"B*01:02:02:01\", \"B*04:01:02\"] },\n}\nground_truth_method = \"method1\"\nresult = compare_method(cohort, ground_truth_method, \"hla\")\n```\n\n#### Print result allele by allele\n``` python\nprint(result)\n\n# Method method2\n# Sample sample_id1\n# A*01:02:03:04 =4= A*01:02:03:04\n# A*01:02 =2= A*01:02:03\n# B*01:01:01:01 =1= B*01:02:02:01\n# B*03:01 =0= B*04:01:02\n# Note:\n# Left hand side is the alleles in reference method/cohort\n# Right hand side is the allele in another method/cohort\n```\n\n\n#### Print summary (i.e. Accuracy vs Resolution, Confusion Matrix)\n\n``` python\n# details are in star_allele_comp/summary.py:print_all_summary\ndf_cohort = result.to_dataframe()\nprint_all_summary(df_cohort)\n```\n\n``` txt\nAccuracy summary\n Accuracy num_match num_ref\nResolution 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN\nmethod\nmethod1 1.0 1.00 1.0 1.0 1.0 0.0 0.0 4 4 4 2 2 0 0 4 4 4 2 2 0 0\nmethod2 1.0 0.75 0.5 0.5 0.5 0.0 0.0 4 3 2 1 1 0 0 4 4 4 2 2 0 0\n\n# Note In the accuracy summary table:\n# * num_match represents the number of alleles that match the alleles in the ground truth method under the specific `Resolution`.\n# * num_ref indicates the number of reference alleles with resolution >= `Resolution`\n# * Accuracy is calculated as the ratio of num_match to num_ref.\n# * Accuracy in FP is False Discovery Rate (FDR)\n# * Accuracy in FN is False Negative Rate (FNR)\n\n\nConfusion matrix (not the same sample)\n Count\n match_res -1 0 1 2 3\n ref_res\n-1 2 0 0 0 0\n 1 1 1 0 0 0\n 2 1 0 2 6 0\n 3 0 0 0 0 1\n 4 0 0 0 0 1\n\n # Note\n # -1 indicates FP or FN\n\n\nAccuracy summary per resolution per gene\n Accuracy num_match num_ref\nResolution 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN 0 1 2 3 4 FP FN\nmethod gene\nmethod1 A 1.0 1.0 1.0 1.0 1.0 0.0 0.0 2 2 2 1 1 0 0 2 2 2 1 1 0 0\n B 1.0 1.0 1.0 1.0 1.0 0.0 0.0 2 2 2 1 1 0 0 2 2 2 1 1 0 0\n```\n\n#### Plot summary (i.e. Accuracy vs Resolution, gene, methods)\n\n``` python\nfigs = plot_summary(df_cohort)\n# You can use Dash to show it\nfrom dash import dcc, html, Dash\napp = Dash(__name__)\napp.layout = html.Div(children=[dcc.Graph(figure=fig) for fig in figs])\napp.run(debug=True)\n```\n\n\n\n## Develop\n\n``` bash\npip install pdoc\npdoc star_allele_comp --docformat google\n```\n\n## Details\n\n\n\n",
"bugtrack_url": null,
"license": "MIT License",
"summary": "Utility to compare star alleles",
"version": "0.2",
"project_urls": {
"Homepage": "https://github.com/linnil1/star_alleles_comparator",
"homepage": "https://github.com/linnil1/star_alleles_comparator",
"repository": "https://github.com/linnil1/star_alleles_comparator"
},
"split_keywords": [
"hla",
"kir",
"comparator",
"allele",
"star-allele"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "c93dba211d03a79223955f3013fd06bb159e217eaa592589cd57ed211262430d",
"md5": "2a8bb12f271f37fffc0c2ff7278fd4e8",
"sha256": "90d112007e18acc4d3b5358092739afc338465f2585008da39e4cffc0211d4a7"
},
"downloads": -1,
"filename": "star_allele_comp-0.2-py3-none-any.whl",
"has_sig": false,
"md5_digest": "2a8bb12f271f37fffc0c2ff7278fd4e8",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10",
"size": 14556,
"upload_time": "2023-08-20T14:26:12",
"upload_time_iso_8601": "2023-08-20T14:26:12.029376Z",
"url": "https://files.pythonhosted.org/packages/c9/3d/ba211d03a79223955f3013fd06bb159e217eaa592589cd57ed211262430d/star_allele_comp-0.2-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "99c4dd100bfb49e23877cdc89e4518e2baba69e290ce8d79928f59c3c090036c",
"md5": "0a03c05567a40f3c8815292dd6b0e432",
"sha256": "2e9e95d69f2f6e60e17548258d3503aa5ed66d7f855763d60380bb000bdc619d"
},
"downloads": -1,
"filename": "star_allele_comp-0.2.tar.gz",
"has_sig": false,
"md5_digest": "0a03c05567a40f3c8815292dd6b0e432",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 15904,
"upload_time": "2023-08-20T14:26:13",
"upload_time_iso_8601": "2023-08-20T14:26:13.162543Z",
"url": "https://files.pythonhosted.org/packages/99/c4/dd100bfb49e23877cdc89e4518e2baba69e290ce8d79928f59c3c090036c/star_allele_comp-0.2.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-08-20 14:26:13",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "linnil1",
"github_project": "star_alleles_comparator",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "star-allele-comp"
}
