
# tdprepview
Python Package that creates Data Preparation Pipelines in Views written in Teradata-SQL.
## Installation
* `pip install tdprepview`
## Features
* Pipeline class that allows creating in-DB preprocessing pipelines
* Several Preprocessor functions
* API similar to sklearn.Pipeline
* Pipeline can be saved and loaded as dict/json
* Pipeline can be automatically created based on data types and distributions.

## Quickstart
### 1. setting up the pipeline
```python
import teradataml as tdml
database_credentials = {
# ...
}
DbCon = tdml.create_context(**database_credentials)
myschema, table_train, table_score = "...", "...", "..."
DF_train = tdml.DataFrame(tdml.in_schema(myschema,table_train))
DF_score = tdml.DataFrame(tdml.in_schema(myschema,table_score))
from tdprepview import Pipeline
from tdprepview import (
StandardScaler, IterativeImputer, PCA, DecisionTreeBinning )
steps = [
({"pattern":"^floatfeat_[1-5]$"}, [StandardScaler(), IterativeImputer()], {"suffix":"_imputed"}),
({"suffix":"_imputed"}, PCA(n_components=2), {"prefix":"mypca_"}),
({"prefix":"mypca_"}, DecisionTreeBinning(target_var="target_class", no_bins=3))
]
mypipeline = Pipeline(steps=steps)
```
### 2.a transform DataFrames directly
this is suitable if you are working in a sandbox Jupyter Notebook
```python
# fit() calculates all necessary statistics in DB
mypipeline.fit(DF=DF_train)
# transform generates SQL syntax based on the chosen Preprocessors and the statistics from fit()
DF_train_transf = mypipeline.transform(DF_train)
DF_score_transf = mypipeline.transform(DF_score)
#... go on with some modelling (e.g. sklearn, TD_DecisionForest,...)
# and scoring (via BYOM, TD_DecisionForestPredict, ...)
```
### 2.b inspect produced queries
if you want to check the SQL code, that is generated py tdprepview
```python
mypipeline.fit(DF=DF_train)
query_train = mypipeline.transform(DF_train, return_type = "str")
print(query_train)
```
the output: (Note how columns not part of the pipeline are simply forwarded)
```sql
WITH preprocessing_steps AS
(
SELECT
row_id AS c_i_35,
floatfeat_1 AS c_i_36,
floatfeat_2 AS c_i_37,
floatfeat_3 AS c_i_38,
target_class AS c_i_39,
ZEROIFNULL( (( c_i_36 ) - -0.2331302 ) / NULLIF( 1.747159 , 0) ) AS c_i_40,
ZEROIFNULL( (( c_i_37 ) - 0.03895576 ) / NULLIF( 1.722347 , 0) ) AS c_i_41,
ZEROIFNULL( (( c_i_38 ) - 0.2363556 ) / NULLIF( 2.808312 , 0) ) AS c_i_42,
-1.118451e-08 + (-0.07714142) * (COALESCE(c_i_41, 1.610355e-09)) + (-0.1758817) * (COALESCE(c_i_42, -4.838372e-09)) AS c_i_43,
4.261288e-09 + (-0.0431946) * (COALESCE(c_i_40, -1.045776e-08)) + (0.6412595) * (COALESCE(c_i_42, -4.838372e-09)) AS c_i_44,
-7.079888e-09 + (-0.118112) * (COALESCE(c_i_40, -1.045776e-08)) + (0.624912) * (COALESCE(c_i_41, 1.610355e-09)) AS c_i_45,
(0.2604757) * (c_i_43) + (-0.681657) * (c_i_44) + (-0.6837369) * (c_i_45) AS c_i_46,
(-0.1047098) * (c_i_43) + (0.6840609) * (c_i_44) + (-0.7218702) * (c_i_45) AS c_i_47,
CASE WHEN c_i_46 < -2.0 THEN 0 WHEN c_i_46 < -1.236351 THEN 1 WHEN c_i_46 < -1.182989 THEN 2 ELSE 3 END AS c_i_48,
CASE WHEN c_i_47 < -2.0 THEN 0 WHEN c_i_47 < -0.3139175 THEN 1 WHEN c_i_47 < 0.2286314 THEN 2 ELSE 3 END AS c_i_49
FROM
<input_schema>.<input_table_view> t
)
SELECT
c_i_35 AS row_id,
c_i_48 AS mypca_pc_1,
c_i_49 AS mypca_pc_2,
c_i_39 AS target_class
FROM
preprocessing_steps t
```
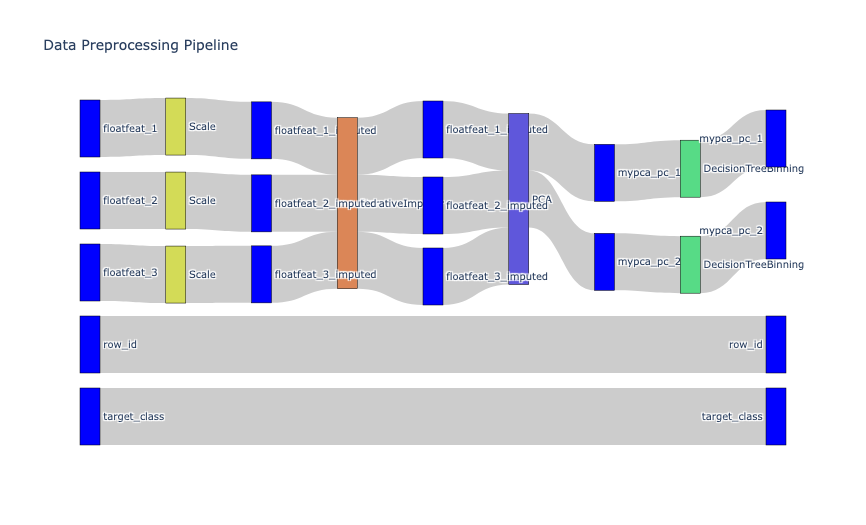
if you want to inspect the pipeline as a chart
```python
mypipeline.plot_sankey()
```
Output:
### 2.c persist transformed data as a view for later use
this is suitable and compliant with the Teradata ModelOps-Framework, where training.py and scoring.py
are separate scripts.
__in `training.py`:__
```python
view_train_transf = table_train+"_transf_v"
view_score_transf = table_score+"_transf_v"
mypipeline.fit(schema_name = myschema,
table_name = table_train)
# 3. transform: create views for pipelines
# DF_train_transf is already based on the newly created View
DF_train_transf = mypipeline.transform(
schema_name = myschema,
table_name = table_train,
# this triggeres the creation of a VIEW, thus the transformation
# pipeline is persited
create_replace_view = True,
output_schema_name = myschema,
output_view_name= view_train_transf)
# 3.b create view for scoring table - no need for further inspection, thus no return
mypipeline.transform( schema_name = myschema,
table_name = table_score,
return_type = None,
create_replace_view = True,
output_schema_name = myschema,
output_view_name= view_score_transf)
# further steps:
# Model training with DF_train_transf,
# e.g. local and use BYOM, or in-DB with TD_DecisionForest
# save model in DB
```
__in `scoring.py`:__
```python
view_score_transf = table_score+"_transf_v"
# 1. get DataFrame based on View, transform is happening as per view definition
DF_score_transf = tdml.DataFrame(tdml.in_schema(myschema, view_score_transf))
# 2. Model Scoring with trained model
# (e.g. PMMLPredict, TD_DecisionForestPredict,...)
# get pointer for model from DB + execute scoring in DB
# 3. Save Scored Data in DB
```
# History
## v0.1.0 (2023-02-15)
### added
* First release on PyPI.
* Pipeline with fit and transform functions
* Preprocessor Functions
* Impute
* ImputeText
* TryCast
* Scale
* CutOff
* FixedWidthBinning
* ThresholdBinarizer
* ListBinarizer
* VariableWidthBinning
* LabelEncoder
* CustomTransformer
* Notebooks for tests
* Demo Notebook
## v0.1.2 (2023-02-15)
### fixed
* added *.sql to MANIFEST.ln such that SQL templates are also part of the distribution.
### changed
* HISTORY and README file from rst to Markdown
## v0.1.3 (2023-02-16)
### added
* Quickstart in README file
## v0.1.4 (2023-02-17)
### added
* DecisionTreeBinning as Preprocessing function
## v1.0.2 (2023-03-06)
Major Overhaul of tdprepview. It now supports transformations that change the schema of the Input DataFrame, like OneHotEncoding. This implementation is based on a directed acyclic graph (DAG).
### added
* plot_sankey() Function for Pipeliine class, which plots the DAG as sankey chart.
* Preprocessing functions
* SimpleImputer (adapted from sklearn, based on Impute)
* IterativeImputer (adapted from sklearn)
* StandardScaler (adapted from sklearn, based on Scale)
* MaxAbsScaler (adapted from sklearn, based on Scale)
* MinMaxScaler (adapted from sklearn, based on Scale)
* RobustScaler (adapted from sklearn, based on Scale)
* Normalizer (adapted from sklearn)
* QuantileTransformer (adapted from sklearn, based on VariableWidthBinning)
* Binarizer (adapted from sklearn, based on ThresholdBinarizer)
* PolynomialFeatures (adapted from sklearn)
* OneHotEncoder (adapted from sklearn)
* PCA (adapted from sklearn)
### changed
* exactly one query is generated, with one WITH AS part, that contains all preprocessing, one final SELECT, that contains all final columns with the correct column names.
* the steps in the steps argument of Pipeline are more flexible. They are either tuples with two elements (input_column(s), preprocessor(s)) or tuples with three elements (input_column(s), preprocessor(s), options)
* input_column(s): string, list of strings or _NEW_ dictionary for dynamic filtering:
* `{'pattern':<regex pattern to search for column names>}`
* `{'prefix':<string column names start with>}`
* `{'suffix':<string column names end with>}`
* `{'dtype_include':<list of tdtypes to include>}`
* `{'dtype_exclude':<list of tdtypes to exclude>}`
* filter conditions can be combined
* preprocessor(s): one or list of Preprocessors.
* options: dict for renaming column names after applying preprocessor. Useful for filtering them in a later step
* `{'prefix':<adds string to beginning of column names>}`
* `{'suffix':<adds string to end of column names>}`
## v1.1.0 (2024-03-01)
bugfixes, compatibility with newer teradataml version, optional requirements, feature hashing
### added
* Preprocessing functions
* SimpleHashEncoder (a text column can be hash encoded using in-DB HASHROW)
### changed
* when a pipeline is crystallised in a view, the package checks the teradataml version and uses either `tdml.get_context().execute(q)` or `tdml.execute_sql(q)`.
* bugfix: `IterativeImputer` query generation
* plotly & seaborn are now optional requirements. Only when `plot_sankey()` is called, it checks whether the packages are available.
## v1.2.0 (2024-03-06)
minor bugfixes, Multi-Label Binarizer
### added
* Preprocessing functions
* MultiLabelBinarizer (a text column with multiple different values separated by a delimiter can be encoded with multiple binary variables)
## v1.3.0 (2024-03-06)
`column_exclude` options for inputs in steps, Cast function
### added
* Preprocessing functions
* Cast (analogous to SQL, useful to convert everything but target and key to float as last step)
## v1.3.1 (2024-03-26)
### added
* Preprocessing functions
* PowerTransformer (analogous to sklearn PowerTransformer)
## v1.3.2 (2024-03-28)
Fitted Pipeline can now be persisted in a serialised file and reused later.
### added
* Pipeline can now be serialized using the
* `mydict = mypipeline.to_dict()` or
* `mypipeline.to_json("mypipeline.json")` functions
* and Pipelines can now analogously be re-created from a serialized representation using the
* `mypipeline = Pipeline.from_dict(mydict)` or
* `mypipeline = Pipeline.from_json("mypipeline.json")` or functions.
## v1.4.0 (2024-04-08)
Introducing automatic Pipeline creation based on heuristics. Either via `Pipeline.from_DataFrame(...)` or via `auto_code(...)`
### added
* Pipeline can now automatically created based on a tdml.DataFrame
* `mypipeline = Pipeline.from_DataFrame(DF, non_feature_cols=["rowid","target"], fit_pipeline=True)`
* It'll use heuristics based on datatypes and distributions to decide which preprocessing function would make sense.
* If you only want to see the code for the `steps` parameter, you can use
* `steps_str = auto_code(DF, non_feature_cols=["rowid","target"])`
* `print(steps_str) # see steps and adjust if needed...`
* `steps = eval(steps_str)`
* `mypipeline = Pipeline(steps)`
## v1.5.0 (2024-09-19)
adding TargetEncoder
### added
* Preprocessing Function
* `TargetEncoder` as in sklearn: Each category is encoded based on a shrunk estimate of the average target values for observations belonging to the category. The encoding scheme mixes the global target mean with the target mean conditioned on the value of the category
Raw data
{
"_id": null,
"home_page": null,
"name": "tdprepview",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": null,
"keywords": "tdprepview, teradata, database, preprocessing, data engineering, data science",
"author": "Martin Hillebrand",
"author_email": "martin.hillebrand@teradata.com",
"download_url": "https://files.pythonhosted.org/packages/e7/c9/1e5c0a7a963000dc65fa7fd5ee2f2772d58cb6eb2c9269334fe16da8575b/tdprepview-1.5.0.tar.gz",
"platform": null,
"description": "\n\n\n# tdprepview\n\nPython Package that creates Data Preparation Pipelines in Views written in Teradata-SQL.\n\n## Installation\n\n\n* `pip install tdprepview`\n\n## Features\n\n* Pipeline class that allows creating in-DB preprocessing pipelines\n* Several Preprocessor functions\n* API similar to sklearn.Pipeline\n* Pipeline can be saved and loaded as dict/json\n* Pipeline can be automatically created based on data types and distributions.\n\n\n\n\n## Quickstart\n\n### 1. setting up the pipeline\n\n```python\nimport teradataml as tdml\ndatabase_credentials = {\n # ...\n}\nDbCon = tdml.create_context(**database_credentials)\n\nmyschema, table_train, table_score = \"...\", \"...\", \"...\"\nDF_train = tdml.DataFrame(tdml.in_schema(myschema,table_train))\nDF_score = tdml.DataFrame(tdml.in_schema(myschema,table_score))\n\nfrom tdprepview import Pipeline\nfrom tdprepview import (\n StandardScaler, IterativeImputer, PCA, DecisionTreeBinning )\n\nsteps = [\n ({\"pattern\":\"^floatfeat_[1-5]$\"}, [StandardScaler(), IterativeImputer()], {\"suffix\":\"_imputed\"}),\n ({\"suffix\":\"_imputed\"}, PCA(n_components=2), {\"prefix\":\"mypca_\"}),\n ({\"prefix\":\"mypca_\"}, DecisionTreeBinning(target_var=\"target_class\", no_bins=3))\n]\n\nmypipeline = Pipeline(steps=steps)\n\n```\n\n### 2.a transform DataFrames directly\nthis is suitable if you are working in a sandbox Jupyter Notebook\n```python\n# fit() calculates all necessary statistics in DB\nmypipeline.fit(DF=DF_train)\n# transform generates SQL syntax based on the chosen Preprocessors and the statistics from fit()\nDF_train_transf = mypipeline.transform(DF_train)\nDF_score_transf = mypipeline.transform(DF_score)\n#... go on with some modelling (e.g. sklearn, TD_DecisionForest,...) \n# and scoring (via BYOM, TD_DecisionForestPredict, ...)\n```\n\n### 2.b inspect produced queries\nif you want to check the SQL code, that is generated py tdprepview\n```python\nmypipeline.fit(DF=DF_train)\nquery_train = mypipeline.transform(DF_train, return_type = \"str\")\nprint(query_train)\n```\nthe output: (Note how columns not part of the pipeline are simply forwarded)\n```sql\nWITH preprocessing_steps AS\n(\n SELECT\n row_id AS c_i_35,\n floatfeat_1 AS c_i_36,\n floatfeat_2 AS c_i_37,\n floatfeat_3 AS c_i_38,\n target_class AS c_i_39,\n ZEROIFNULL( (( c_i_36 ) - -0.2331302 ) / NULLIF( 1.747159 , 0) ) AS c_i_40,\n ZEROIFNULL( (( c_i_37 ) - 0.03895576 ) / NULLIF( 1.722347 , 0) ) AS c_i_41,\n ZEROIFNULL( (( c_i_38 ) - 0.2363556 ) / NULLIF( 2.808312 , 0) ) AS c_i_42,\n -1.118451e-08 + (-0.07714142) * (COALESCE(c_i_41, 1.610355e-09)) + (-0.1758817) * (COALESCE(c_i_42, -4.838372e-09)) AS c_i_43,\n 4.261288e-09 + (-0.0431946) * (COALESCE(c_i_40, -1.045776e-08)) + (0.6412595) * (COALESCE(c_i_42, -4.838372e-09)) AS c_i_44,\n -7.079888e-09 + (-0.118112) * (COALESCE(c_i_40, -1.045776e-08)) + (0.624912) * (COALESCE(c_i_41, 1.610355e-09)) AS c_i_45,\n (0.2604757) * (c_i_43) + (-0.681657) * (c_i_44) + (-0.6837369) * (c_i_45) AS c_i_46,\n (-0.1047098) * (c_i_43) + (0.6840609) * (c_i_44) + (-0.7218702) * (c_i_45) AS c_i_47,\n CASE WHEN c_i_46 < -2.0 THEN 0 WHEN c_i_46 < -1.236351 THEN 1 WHEN c_i_46 < -1.182989 THEN 2 ELSE 3 END AS c_i_48,\n CASE WHEN c_i_47 < -2.0 THEN 0 WHEN c_i_47 < -0.3139175 THEN 1 WHEN c_i_47 < 0.2286314 THEN 2 ELSE 3 END AS c_i_49\n FROM\n <input_schema>.<input_table_view> t\n)\n\nSELECT\n c_i_35 AS row_id,\n c_i_48 AS mypca_pc_1,\n c_i_49 AS mypca_pc_2,\n c_i_39 AS target_class\nFROM\npreprocessing_steps t\n```\n\nif you want to inspect the pipeline as a chart\n```python\nmypipeline.plot_sankey()\n```\nOutput:\n\n\n\n\n\n### 2.c persist transformed data as a view for later use\nthis is suitable and compliant with the Teradata ModelOps-Framework, where training.py and scoring.py \nare separate scripts.\n\n__in `training.py`:__\n```python\nview_train_transf = table_train+\"_transf_v\"\nview_score_transf = table_score+\"_transf_v\"\n\nmypipeline.fit(schema_name = myschema, \n table_name = table_train)\n\n# 3. transform: create views for pipelines\n# DF_train_transf is already based on the newly created View\nDF_train_transf = mypipeline.transform(\n schema_name = myschema, \n table_name = table_train,\n # this triggeres the creation of a VIEW, thus the transformation \n # pipeline is persited\n create_replace_view = True, \n output_schema_name = myschema, \n output_view_name= view_train_transf)\n\n# 3.b create view for scoring table - no need for further inspection, thus no return\nmypipeline.transform( schema_name = myschema, \n table_name = table_score,\n return_type = None,\n create_replace_view = True, \n output_schema_name = myschema, \n output_view_name= view_score_transf)\n\n# further steps:\n# Model training with DF_train_transf, \n# e.g. local and use BYOM, or in-DB with TD_DecisionForest\n# save model in DB\n```\n\n__in `scoring.py`:__\n```python\nview_score_transf = table_score+\"_transf_v\"\n# 1. get DataFrame based on View, transform is happening as per view definition\nDF_score_transf = tdml.DataFrame(tdml.in_schema(myschema, view_score_transf))\n\n# 2. Model Scoring with trained model \n# (e.g. PMMLPredict, TD_DecisionForestPredict,...)\n# get pointer for model from DB + execute scoring in DB\n\n# 3. Save Scored Data in DB\n```\n\n\n\n# History\n\n## v0.1.0 (2023-02-15)\n\n### added\n\n* First release on PyPI.\n* Pipeline with fit and transform functions\n* Preprocessor Functions\n * Impute\n * ImputeText\n * TryCast\n * Scale\n * CutOff\n * FixedWidthBinning\n * ThresholdBinarizer\n * ListBinarizer\n * VariableWidthBinning\n * LabelEncoder\n * CustomTransformer\n* Notebooks for tests\n* Demo Notebook\n\n## v0.1.2 (2023-02-15)\n\n### fixed\n\n* added *.sql to MANIFEST.ln such that SQL templates are also part of the distribution.\n\n### changed\n\n* HISTORY and README file from rst to Markdown\n\n## v0.1.3 (2023-02-16)\n\n### added\n\n* Quickstart in README file\n\n## v0.1.4 (2023-02-17)\n\n### added\n\n* DecisionTreeBinning as Preprocessing function\n\n## v1.0.2 (2023-03-06)\n\nMajor Overhaul of tdprepview. It now supports transformations that change the schema of the Input DataFrame, like OneHotEncoding. This implementation is based on a directed acyclic graph (DAG). \n\n### added\n\n* plot_sankey() Function for Pipeliine class, which plots the DAG as sankey chart.\n* Preprocessing functions\n * SimpleImputer (adapted from sklearn, based on Impute)\n * IterativeImputer (adapted from sklearn)\n * StandardScaler (adapted from sklearn, based on Scale)\n * MaxAbsScaler (adapted from sklearn, based on Scale)\n * MinMaxScaler (adapted from sklearn, based on Scale)\n * RobustScaler (adapted from sklearn, based on Scale)\n * Normalizer (adapted from sklearn)\n * QuantileTransformer (adapted from sklearn, based on VariableWidthBinning)\n * Binarizer (adapted from sklearn, based on ThresholdBinarizer)\n * PolynomialFeatures (adapted from sklearn)\n * OneHotEncoder (adapted from sklearn)\n * PCA (adapted from sklearn)\n\n### changed\n\n* exactly one query is generated, with one WITH AS part, that contains all preprocessing, one final SELECT, that contains all final columns with the correct column names.\n* the steps in the steps argument of Pipeline are more flexible. They are either tuples with two elements (input_column(s), preprocessor(s)) or tuples with three elements (input_column(s), preprocessor(s), options)\n * input_column(s): string, list of strings or _NEW_ dictionary for dynamic filtering:\n * `{'pattern':<regex pattern to search for column names>}`\n * `{'prefix':<string column names start with>}`\n * `{'suffix':<string column names end with>}`\n * `{'dtype_include':<list of tdtypes to include>}`\n * `{'dtype_exclude':<list of tdtypes to exclude>}`\n * filter conditions can be combined\n * preprocessor(s): one or list of Preprocessors.\n * options: dict for renaming column names after applying preprocessor. Useful for filtering them in a later step\n * `{'prefix':<adds string to beginning of column names>}`\n * `{'suffix':<adds string to end of column names>}`\n\n## v1.1.0 (2024-03-01)\n\nbugfixes, compatibility with newer teradataml version, optional requirements, feature hashing\n\n### added\n\n* Preprocessing functions\n * SimpleHashEncoder (a text column can be hash encoded using in-DB HASHROW)\n\n### changed\n\n* when a pipeline is crystallised in a view, the package checks the teradataml version and uses either `tdml.get_context().execute(q)` or `tdml.execute_sql(q)`.\n* bugfix: `IterativeImputer` query generation\n* plotly & seaborn are now optional requirements. Only when `plot_sankey()` is called, it checks whether the packages are available.\n\n\n\n## v1.2.0 (2024-03-06)\n\nminor bugfixes, Multi-Label Binarizer\n\n### added\n\n* Preprocessing functions\n * MultiLabelBinarizer (a text column with multiple different values separated by a delimiter can be encoded with multiple binary variables)\n\n\n## v1.3.0 (2024-03-06)\n\n`column_exclude` options for inputs in steps, Cast function\n\n### added\n\n* Preprocessing functions\n * Cast (analogous to SQL, useful to convert everything but target and key to float as last step)\n\n\n## v1.3.1 (2024-03-26)\n\n### added\n\n* Preprocessing functions\n * PowerTransformer (analogous to sklearn PowerTransformer)\n\n## v1.3.2 (2024-03-28)\n\nFitted Pipeline can now be persisted in a serialised file and reused later.\n\n### added\n\n* Pipeline can now be serialized using the \n * `mydict = mypipeline.to_dict()` or \n * `mypipeline.to_json(\"mypipeline.json\")` functions\n* and Pipelines can now analogously be re-created from a serialized representation using the\n * `mypipeline = Pipeline.from_dict(mydict)` or\n * `mypipeline = Pipeline.from_json(\"mypipeline.json\")` or functions.\n\n## v1.4.0 (2024-04-08)\n\nIntroducing automatic Pipeline creation based on heuristics. Either via `Pipeline.from_DataFrame(...)` or via `auto_code(...)`\n\n### added\n\n* Pipeline can now automatically created based on a tdml.DataFrame \n * `mypipeline = Pipeline.from_DataFrame(DF, non_feature_cols=[\"rowid\",\"target\"], fit_pipeline=True)`\n * It'll use heuristics based on datatypes and distributions to decide which preprocessing function would make sense.\n* If you only want to see the code for the `steps` parameter, you can use\n * `steps_str = auto_code(DF, non_feature_cols=[\"rowid\",\"target\"])`\n * `print(steps_str) # see steps and adjust if needed...` \n * `steps = eval(steps_str)`\n * `mypipeline = Pipeline(steps)`\n\n## v1.5.0 (2024-09-19)\n\nadding TargetEncoder\n\n### added\n\n* Preprocessing Function\n * `TargetEncoder` as in sklearn: Each category is encoded based on a shrunk estimate of the average target values for observations belonging to the category. The encoding scheme mixes the global target mean with the target mean conditioned on the value of the category\n",
"bugtrack_url": null,
"license": null,
"summary": "Python Package that creates Data Preparation Pipeline in Teradata-SQL in Views",
"version": "1.5.0",
"project_urls": null,
"split_keywords": [
"tdprepview",
" teradata",
" database",
" preprocessing",
" data engineering",
" data science"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "89ad64548631bfcf9c01a9f75b0733114d56362b475b663531da31c75629e55b",
"md5": "9c211205d636c65e83c244723aec4fdd",
"sha256": "0aa1abf8b1b61bbedf093e3b55a38b5750d80ff68551761a9939a99fb3919fa0"
},
"downloads": -1,
"filename": "tdprepview-1.5.0-py2.py3-none-any.whl",
"has_sig": false,
"md5_digest": "9c211205d636c65e83c244723aec4fdd",
"packagetype": "bdist_wheel",
"python_version": "py2.py3",
"requires_python": ">=3.8",
"size": 72225,
"upload_time": "2024-09-23T16:04:59",
"upload_time_iso_8601": "2024-09-23T16:04:59.828402Z",
"url": "https://files.pythonhosted.org/packages/89/ad/64548631bfcf9c01a9f75b0733114d56362b475b663531da31c75629e55b/tdprepview-1.5.0-py2.py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "e7c91e5c0a7a963000dc65fa7fd5ee2f2772d58cb6eb2c9269334fe16da8575b",
"md5": "206bfdd131c13d4f118fa8a1254e8c04",
"sha256": "569a05582035bd4f25bfd122280f74a01273d938acfe1ce56abeedd1afd90d91"
},
"downloads": -1,
"filename": "tdprepview-1.5.0.tar.gz",
"has_sig": false,
"md5_digest": "206bfdd131c13d4f118fa8a1254e8c04",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 2034612,
"upload_time": "2024-09-23T16:05:03",
"upload_time_iso_8601": "2024-09-23T16:05:03.004903Z",
"url": "https://files.pythonhosted.org/packages/e7/c9/1e5c0a7a963000dc65fa7fd5ee2f2772d58cb6eb2c9269334fe16da8575b/tdprepview-1.5.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-09-23 16:05:03",
"github": false,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"lcname": "tdprepview"
}