# threshold-clustering
## Threshold Spectral Community Detection for NetworkX
NetworkX Community detection based on the algorithm proposed in Guzzi et. al. 2013 (*).
Developed for semantic similarity networks, this algorithm specifically targets weighted and directed graphs.
This implementation adds a couple of options to the algorithm proposed in the paper, such as passing an arbitrary community detection function (e.g. python-louvain).
Similarity networks are typically dense, weighted and difficult to cluster. Experience shows that algorithms such as python-louvain
have difficulty finding outliers and smaller partitions.
Given a networkX.DiGraph object, threshold-clustering will try to remove insignificant ties according to a local threshold.
This threshold is refined until the network breaks into distinct components in a sparse, undirected network.
As a next step, either these components are taken communities directly, or, alternatively, another community detection (e.g. python-louvain)
can be applied.
## Example
Consider the cosine similarities in the Karate Club Network. Although these similarities are not directed, they are rather dense.
```python
import networkx as nx
import numpy as np
import matplotlib.cm as cm
import matplotlib.pyplot as plt
from sklearn.metrics.pairwise import cosine_similarity
# load graph
G = nx.karate_club_graph()
# Generate a similarity style weighted graph
Adj=nx.to_numpy_matrix(G)
cos_Adj=cosine_similarity(Adj.T)
G=nx.from_numpy_matrix(cos_Adj)
pos = nx.spring_layout(G)
weights = np.array([G[u][v]['weight'] for u,v in G.edges()])*5
nx.draw_networkx_nodes(G, pos, node_size=40)
nx.draw_networkx_edges(G, pos, alpha=0.2, width=weights)
plt.show()
```
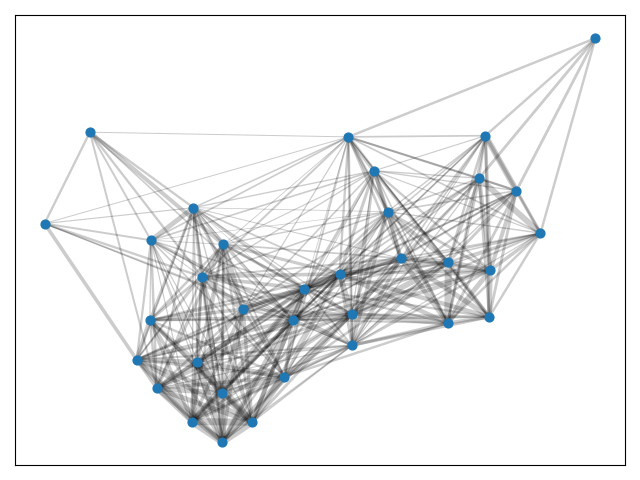
Let's use python-louvain to find the best partition.
```python
partition=community_louvain.best_partition(G.to_undirected())
cmap = cm.get_cmap('viridis', max(partition.values()) + 1)
nx.draw_networkx_nodes(G, pos, partition.keys(), node_size=40,
cmap=cmap, node_color=list(partition.values()))
nx.draw_networkx_edges(G, pos, alpha=0.2,width=weights)
plt.show()
```
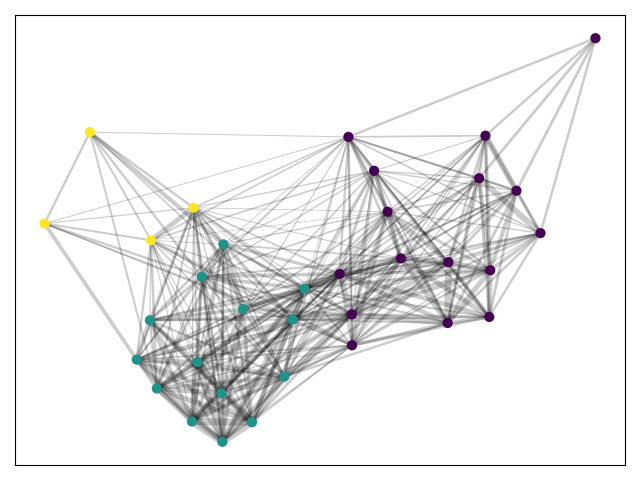
We get three rather large partition and no sense of outliers.
Instead, we can use threshold-clustering's best_partition function to run python_louvain's community detection on a
transformed network.
```python
from thresholdclustering import best_partition
cluster_function = community_louvain.best_partition
partition, alpha = best_partition(G, cluster_function=cluster_function)
cmap = cm.get_cmap('viridis', max(partition.values()) + 1)
nx.draw_networkx_nodes(G, pos, partition.keys(), node_size=40,
cmap=cmap, node_color=list(partition.values()))
nx.draw_networkx_edges(G, pos, alpha=0.2,width=weights)
plt.show()
```
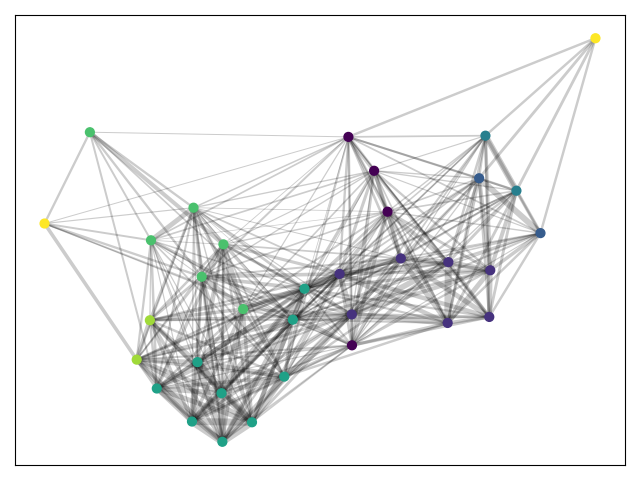
We can see that more communities of similarity can be identified. Note in particular outliers drawn in yellow.
## Installation
`(*) Guzzi, Pietro Hiram, Pierangelo Veltri, and Mario Cannataro. "Thresholding of semantic similarity networks using a spectral graph-based technique."
International Workshop on New Frontiers in Mining Complex Patterns. Springer, Cham, 2013.`
# History
## 1.0
First release. Since this is a small, yet nifty add-on to python-louvain, it is a quick write.
Please let me know if anything more should be added!
Raw data
{
"_id": null,
"home_page": "https://github.com/IngoMarquart/python-threshold-clustering",
"name": "thresholdclustering",
"maintainer": "",
"docs_url": null,
"requires_python": "",
"maintainer_email": "",
"keywords": "community detection,clustering,networkx,python-louvain",
"author": "Ingo Marquart",
"author_email": "ingo.marquart@esmt.org",
"download_url": "https://files.pythonhosted.org/packages/27/38/84c0b0662503d94cb09a5709447257fa9b8ecc93648618d56d7303943cdd/thresholdclustering-1.1.tar.gz",
"platform": "",
"description": "# threshold-clustering\n\n## Threshold Spectral Community Detection for NetworkX\n\n\nNetworkX Community detection based on the algorithm proposed in Guzzi et. al. 2013 (*).\n\nDeveloped for semantic similarity networks, this algorithm specifically targets weighted and directed graphs. \nThis implementation adds a couple of options to the algorithm proposed in the paper, such as passing an arbitrary community detection function (e.g. python-louvain).\n\nSimilarity networks are typically dense, weighted and difficult to cluster. Experience shows that algorithms such as python-louvain \nhave difficulty finding outliers and smaller partitions.\n\nGiven a networkX.DiGraph object, threshold-clustering will try to remove insignificant ties according to a local threshold.\nThis threshold is refined until the network breaks into distinct components in a sparse, undirected network.\n\nAs a next step, either these components are taken communities directly, or, alternatively, another community detection (e.g. python-louvain)\ncan be applied.\n\n\n## Example\n\nConsider the cosine similarities in the Karate Club Network. Although these similarities are not directed, they are rather dense.\n\n```python\nimport networkx as nx\nimport numpy as np\nimport matplotlib.cm as cm\nimport matplotlib.pyplot as plt\nfrom sklearn.metrics.pairwise import cosine_similarity\n\n# load graph\nG = nx.karate_club_graph()\n\n# Generate a similarity style weighted graph\nAdj=nx.to_numpy_matrix(G)\ncos_Adj=cosine_similarity(Adj.T)\nG=nx.from_numpy_matrix(cos_Adj)\n\npos = nx.spring_layout(G)\nweights = np.array([G[u][v]['weight'] for u,v in G.edges()])*5\nnx.draw_networkx_nodes(G, pos, node_size=40)\nnx.draw_networkx_edges(G, pos, alpha=0.2, width=weights)\nplt.show()\n```\n\n\n\n\nLet's use python-louvain to find the best partition.\n\n```python\npartition=community_louvain.best_partition(G.to_undirected())\n\ncmap = cm.get_cmap('viridis', max(partition.values()) + 1)\nnx.draw_networkx_nodes(G, pos, partition.keys(), node_size=40,\n cmap=cmap, node_color=list(partition.values()))\nnx.draw_networkx_edges(G, pos, alpha=0.2,width=weights)\nplt.show()\n```\n\n\n\nWe get three rather large partition and no sense of outliers.\n\nInstead, we can use threshold-clustering's best_partition function to run python_louvain's community detection on a\ntransformed network. \n\n\n```python\nfrom thresholdclustering import best_partition\n\ncluster_function = community_louvain.best_partition\npartition, alpha = best_partition(G, cluster_function=cluster_function)\n\n\n\ncmap = cm.get_cmap('viridis', max(partition.values()) + 1)\nnx.draw_networkx_nodes(G, pos, partition.keys(), node_size=40,\n cmap=cmap, node_color=list(partition.values()))\nnx.draw_networkx_edges(G, pos, alpha=0.2,width=weights)\nplt.show()\n```\n\n\n\n\n\nWe can see that more communities of similarity can be identified. Note in particular outliers drawn in yellow.\n\n\n## Installation\n\n\n\n`(*) Guzzi, Pietro Hiram, Pierangelo Veltri, and Mario Cannataro. \"Thresholding of semantic similarity networks using a spectral graph-based technique.\" \nInternational Workshop on New Frontiers in Mining Complex Patterns. Springer, Cham, 2013.`\n\n\n\n\n# History\n\n## 1.0\n\nFirst release. Since this is a small, yet nifty add-on to python-louvain, it is a quick write.\nPlease let me know if anything more should be added!\n\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Community detection for directed, weighted networkX graphs with spectral thresholding.",
"version": "1.1",
"project_urls": {
"Download": "https://pypi.org/project/thresholdclustering",
"Homepage": "https://github.com/IngoMarquart/python-threshold-clustering"
},
"split_keywords": [
"community detection",
"clustering",
"networkx",
"python-louvain"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "49262b0d326e9d637c687a8f1aeeb5b7414866576a6721355bdf6532d05d4793",
"md5": "295c2d6559062e24def9e5dc89d77786",
"sha256": "fdad9cd6f58b1e551b0232f3d76f662622aaaacb27397a0372ebb8f9cd629e10"
},
"downloads": -1,
"filename": "thresholdclustering-1.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "295c2d6559062e24def9e5dc89d77786",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": null,
"size": 5304,
"upload_time": "2021-03-06T19:02:36",
"upload_time_iso_8601": "2021-03-06T19:02:36.387436Z",
"url": "https://files.pythonhosted.org/packages/49/26/2b0d326e9d637c687a8f1aeeb5b7414866576a6721355bdf6532d05d4793/thresholdclustering-1.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "273884c0b0662503d94cb09a5709447257fa9b8ecc93648618d56d7303943cdd",
"md5": "f601510e98a635957d336572e8133e70",
"sha256": "af27ceec7a4a8903e1b34e0d57ac28a42deb7d2beff3c403330579b11525bee1"
},
"downloads": -1,
"filename": "thresholdclustering-1.1.tar.gz",
"has_sig": false,
"md5_digest": "f601510e98a635957d336572e8133e70",
"packagetype": "sdist",
"python_version": "source",
"requires_python": null,
"size": 4190,
"upload_time": "2021-03-06T19:02:37",
"upload_time_iso_8601": "2021-03-06T19:02:37.634401Z",
"url": "https://files.pythonhosted.org/packages/27/38/84c0b0662503d94cb09a5709447257fa9b8ecc93648618d56d7303943cdd/thresholdclustering-1.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2021-03-06 19:02:37",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "IngoMarquart",
"github_project": "python-threshold-clustering",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "thresholdclustering"
}
