## Python package for working with the AAindex database (https://www.genome.jp/aaindex/) <a name="TOP"></a>
<p align="center">
<img src="https://raw.githubusercontent.com/amckenna41/aaindex/main/images/aaindex_logo.png" />
</p>
[](https://pypi.org/project/aaindex/)
[](https://github.com/amckenna41/aaindex/actions?query=workflowBuilding%20and%20Testing)
[](https://dl.circleci.com/status-badge/redirect/gh/amckenna41/aaindex/tree/main)
[](https://pypi.org/project/aaindex/)
[](https://pypi.org/project/aaindex/)
[](https://opensource.org/licenses/MIT)
[](https://github.com/amckenna41/aaindex/issues)
<!-- [](https://github.com/amckenna41/aaindex) -->
<!-- [](https://codecov.io/gh/amckenna41/aaindex) -->
Table of Contents
-----------------
* [Introduction](#introduction)
* [Requirements](#requirements)
* [Installation](#installation)
* [Usage](#usage)
* [Tests](#tests)
* [Contact](#contact)
* [License](#license)
* [References](#References)
Introduction
------------
The AAindex is a database of numerical indices representing various physicochemical, structural and biochemical properties of amino acids and pairs of amino acids 🧬. The AAindex consists of three sections: AAindex1 for the amino acid index of 20 numerical values, AAindex2 for the amino acid mutation matrix and AAindex3 for the statistical protein contact potentials. All data are derived from published literature [[1]](#references).
This `aaindex` Python software package is a very lightweight way of accessing the data represented in the various AAindex databases, requiring no additional external library installations. Any record within the 3 databases and their associated data/numerical indices can be accessed in one simple command. Currently the software supports the AAindex1 database with plans to include the AAindex 2 & 3 in the future.
<strong>A demo of the software is available [here](https://colab.research.google.com/drive/1dccV_n1BRMiU8W13F9PPXbSaFzvOdQLC?usp=sharing). </strong>
<!-- ### Format of AAindex1 record
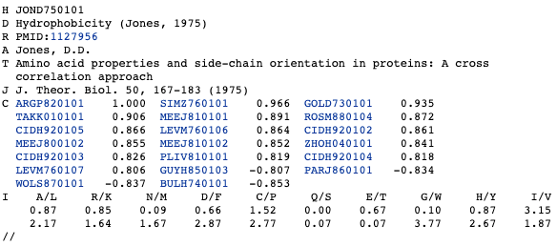
```
************************************************************************
* *
* H Accession number *
* D Data description *
* R Pub med article ID (PMID) *
* A Author(s) *
* T Title of the article *
* J Journal reference *
* * Comment or missing *
* C Accession numbers of similar entries with the correlation *
* coefficients of 0.8 (-0.8) or more (less). *
* Notice: The correlation coefficient is calculated with zeros *
* filled for missing values. *
* I Amino acid index data in the following order *
* Ala Arg Asn Asp Cys Gln Glu Gly His Ile *
* Leu Lys Met Phe Pro Ser Thr Trp Tyr Val *
* // *
************************************************************************
``` -->
Installation
-----------------
Install the latest version of `aaindex` using pip:
```bash
pip3 install aaindex --upgrade
```
Install by cloning repository:
```bash
git clone https://github.com/amckenna41/aaindex.git
python3 setup.py install
```
Usage
-----
The AAindex module is made up of three modules for each AAindex database, with each having a Python class of the same name, when importing the package you should import the required database module:
```python
from aaindex import aaindex1
# from aaindex import aaindex2
# from aaindex import aaindex3
```
## AAIndex1 Usage
### Get record from AAindex1
The AAindex1 class offers diverse functionalities for obtaining any element from any record in the database. The records are imported from a parsed json in the data folder of the package. You can search for a particular record by its record code/accession number or its name/description. You can also get the record category, references, notes, correlation coefficients, pmid and importantly its associated amino acid values:
```python
from aaindex import aaindex1
full_record = aaindex1['CHOP780206'] #get full AAI record
''' full_record ->
{'category': 'sec_struct', 'correlation_coefficients': {}, 'description': 'Normalized frequency of N-terminal non helical region (Chou-Fasman, 1978b)', 'notes': '', 'pmid': '364941', 'references': "Chou, P.Y. and Fasman, G.D. 'Prediction of the secondary structure of proteins from their amino acid sequence' Adv. Enzymol. 47, 45-148 (1978)", 'values': {'-': 0, 'A': 0.7, 'C': 0.65, 'D': 0.98, 'E': 1.04, 'F': 0.93, 'G': 1.41, 'H': 1.22, 'I': 0.78, 'K': 1.01, 'L': 0.85, 'M': 0.83, 'N': 1.42, 'P': 1.1, 'Q': 0.75, 'R': 0.34, 'S': 1.55, 'T': 1.09, 'V': 0.75, 'W': 0.62, 'Y': 0.99}}
'''
#get individual elements of AAindex record
record_values = aaindex1['CHOP780206']['values']
record_values = aaindex1['CHOP780206'].values
#'values': {'-': 0, 'A': 0.7, 'C': 0.65, 'D': 0.98, 'E': 1.04, 'F': 0.93, 'G': 1.41, 'H': 1.22, 'I': 0.78, 'K': 1.01, 'L': 0.85, 'M': 0.83, 'N': 1.42, 'P': 1.1, 'Q': 0.75, 'R': 0.34, 'S': 1.55, 'T': 1.09, 'V': 0.75, 'W': 0.62, 'Y': 0.99}
record_description = aaindex1['CHOP780206']['description']
record_description = aaindex1['CHOP780206'].description
#'description': 'Normalized frequency of N-terminal non helical region (Chou-Fasman, 1978b)'
record_references = aaindex1['CHOP780206']['references']
record_references = aaindex1['CHOP780206'].references
#'references': "Chou, P.Y. and Fasman, G.D. 'Prediction of the secondary structure of proteins from their amino acid sequence' Adv. Enzymol. 47, 45-148 (1978)"
record_notes = aaindex1['CHOP780206']['notes']
record_notes = aaindex1['CHOP780206'].notes
#""
record_correlation_coefficient = aaindex1['CHOP780206']['correlation_coefficient']
record_correlation_coefficient = aaindex1['CHOP780206'].correlation_coefficient
#{}
record_pmid = aaindex1['CHOP780206']['pmid']
record_pmid = aaindex1['CHOP780206'].pmid
#364941
record_category = aaindex1['CHOP780206']['category']
record_category = aaindex1['CHOP780206'].category
#sec_struct
```
### Get total number of AAindex1 records
```python
#get total number of records in AAI database
aaindex1.num_records()
```
### Get list of all AAindex1 record names
```python
#get list of all AAindex1 record names
aaindex1.record_names()
```
## AAIndex2 Usage
```python
# from aaindex import aaindex1
from aaindex import aaindex2
# from aaindex import aaindex3
```
## AAIndex3 Usage
```python
# from aaindex import aaindex1
# from aaindex import aaindex2
from aaindex import aaindex3
```
Directories 📁
--------------
* `/tests` - unit and integration tests for `aaindex` package.
* `/aaindex` - source code and all required external data files for package.
* `/images` - images used throughout README.
* `/docs` - `aaindex` documentation.
Tests 🧪
--------
To run all tests, from the main `aaindex` folder run:
```
python3 -m unittest discover tests
```
Contact
-------
If you have any questions or comments, please contact amckenna41@qub.ac.uk or raise an issue on the [Issues][Issues] tab.
License
-------
Distributed under the MIT License. See `LICENSE` for more details.
References
----------
\[1\]: Shuichi Kawashima, Minoru Kanehisa, AAindex: Amino Acid index database, Nucleic Acids Research, Volume 28, Issue 1, 1 January 2000, Page 374, https://doi.org/10.1093/nar/28.1.374 <br>
\[2\]: https://www.genome.jp/aaindex/
[Back to top](#TOP)
[python]: https://www.python.org/downloads/release/python-360/
[aaindex]: https://github.com/amckenna41/aaindex
[requests]: https://requests.readthedocs.io/en/latest/
[numpy]: https://numpy.org/
[PyPi]: https://pypi.org/project/aaindex/
[demo]: https://colab.research.google.com/drive/1dccV_n1BRMiU8W13F9PPXbSaFzvOdQLC?usp=sharing
[Issues]: https://github.com/amckenna41/aaindex/issues
Raw data
{
"_id": null,
"home_page": "https://github.com/amckenna41/aaindex",
"name": "aaindex",
"maintainer": "AJ McKenna",
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": "",
"keywords": "amino acid index,aaindex,bioinformatics,protein engineering,python,pypi,physiochemical properties,biochemical properties,proteins,protein structure prediction,pysar",
"author": "AJ McKenna, https://github.com/amckenna41",
"author_email": "amckenna41@qub.ac.uk",
"download_url": "https://github.com/amckenna41/aaindex/archive/refs/heads/main.zip",
"platform": null,
"description": "## Python package for working with the AAindex database (https://www.genome.jp/aaindex/) <a name=\"TOP\"></a>\n<p align=\"center\">\n <img src=\"https://raw.githubusercontent.com/amckenna41/aaindex/main/images/aaindex_logo.png\" />\n</p>\n\n[](https://pypi.org/project/aaindex/)\n[](https://github.com/amckenna41/aaindex/actions?query=workflowBuilding%20and%20Testing)\n[](https://dl.circleci.com/status-badge/redirect/gh/amckenna41/aaindex/tree/main)\n[](https://pypi.org/project/aaindex/)\n[](https://pypi.org/project/aaindex/)\n[](https://opensource.org/licenses/MIT)\n[](https://github.com/amckenna41/aaindex/issues)\n<!-- [](https://github.com/amckenna41/aaindex) -->\n<!-- [](https://codecov.io/gh/amckenna41/aaindex) -->\n\nTable of Contents\n-----------------\n\n * [Introduction](#introduction)\n * [Requirements](#requirements)\n * [Installation](#installation)\n * [Usage](#usage)\n * [Tests](#tests)\n * [Contact](#contact)\n * [License](#license)\n * [References](#References)\n\nIntroduction\n------------\nThe AAindex is a database of numerical indices representing various physicochemical, structural and biochemical properties of amino acids and pairs of amino acids \ud83e\uddec. The AAindex consists of three sections: AAindex1 for the amino acid index of 20 numerical values, AAindex2 for the amino acid mutation matrix and AAindex3 for the statistical protein contact potentials. All data are derived from published literature [[1]](#references). \n\nThis `aaindex` Python software package is a very lightweight way of accessing the data represented in the various AAindex databases, requiring no additional external library installations. Any record within the 3 databases and their associated data/numerical indices can be accessed in one simple command. Currently the software supports the AAindex1 database with plans to include the AAindex 2 & 3 in the future.\n\n<strong>A demo of the software is available [here](https://colab.research.google.com/drive/1dccV_n1BRMiU8W13F9PPXbSaFzvOdQLC?usp=sharing). </strong>\n\n<!-- ### Format of AAindex1 record\n\n\n```\n ************************************************************************\n * *\n * H Accession number *\n * D Data description *\n * R Pub med article ID (PMID) *\n * A Author(s) *\n * T Title of the article *\n * J Journal reference *\n * * Comment or missing *\n * C Accession numbers of similar entries with the correlation *\n * coefficients of 0.8 (-0.8) or more (less). *\n * Notice: The correlation coefficient is calculated with zeros *\n * filled for missing values. *\n * I Amino acid index data in the following order *\n * Ala Arg Asn Asp Cys Gln Glu Gly His Ile *\n * Leu Lys Met Phe Pro Ser Thr Trp Tyr Val *\n * // *\n ************************************************************************\n``` -->\nInstallation\n-----------------\nInstall the latest version of `aaindex` using pip:\n\n```bash\npip3 install aaindex --upgrade\n```\n\nInstall by cloning repository:\n\n```bash\ngit clone https://github.com/amckenna41/aaindex.git\npython3 setup.py install\n```\nUsage\n-----\nThe AAindex module is made up of three modules for each AAindex database, with each having a Python class of the same name, when importing the package you should import the required database module:\n\n```python\nfrom aaindex import aaindex1\n# from aaindex import aaindex2\n# from aaindex import aaindex3\n```\n\n## AAIndex1 Usage\n\n### Get record from AAindex1\nThe AAindex1 class offers diverse functionalities for obtaining any element from any record in the database. The records are imported from a parsed json in the data folder of the package. You can search for a particular record by its record code/accession number or its name/description. You can also get the record category, references, notes, correlation coefficients, pmid and importantly its associated amino acid values:\n```python\nfrom aaindex import aaindex1\n\nfull_record = aaindex1['CHOP780206'] #get full AAI record\n''' full_record ->\n{'category': 'sec_struct', 'correlation_coefficients': {}, 'description': 'Normalized frequency of N-terminal non helical region (Chou-Fasman, 1978b)', 'notes': '', 'pmid': '364941', 'references': \"Chou, P.Y. and Fasman, G.D. 'Prediction of the secondary structure of proteins from their amino acid sequence' Adv. Enzymol. 47, 45-148 (1978)\", 'values': {'-': 0, 'A': 0.7, 'C': 0.65, 'D': 0.98, 'E': 1.04, 'F': 0.93, 'G': 1.41, 'H': 1.22, 'I': 0.78, 'K': 1.01, 'L': 0.85, 'M': 0.83, 'N': 1.42, 'P': 1.1, 'Q': 0.75, 'R': 0.34, 'S': 1.55, 'T': 1.09, 'V': 0.75, 'W': 0.62, 'Y': 0.99}}\n'''\n\n#get individual elements of AAindex record\nrecord_values = aaindex1['CHOP780206']['values'] \nrecord_values = aaindex1['CHOP780206'].values\n#'values': {'-': 0, 'A': 0.7, 'C': 0.65, 'D': 0.98, 'E': 1.04, 'F': 0.93, 'G': 1.41, 'H': 1.22, 'I': 0.78, 'K': 1.01, 'L': 0.85, 'M': 0.83, 'N': 1.42, 'P': 1.1, 'Q': 0.75, 'R': 0.34, 'S': 1.55, 'T': 1.09, 'V': 0.75, 'W': 0.62, 'Y': 0.99}\n\nrecord_description = aaindex1['CHOP780206']['description']\nrecord_description = aaindex1['CHOP780206'].description\n#'description': 'Normalized frequency of N-terminal non helical region (Chou-Fasman, 1978b)'\n\nrecord_references = aaindex1['CHOP780206']['references']\nrecord_references = aaindex1['CHOP780206'].references\n#'references': \"Chou, P.Y. and Fasman, G.D. 'Prediction of the secondary structure of proteins from their amino acid sequence' Adv. Enzymol. 47, 45-148 (1978)\"\n\nrecord_notes = aaindex1['CHOP780206']['notes']\nrecord_notes = aaindex1['CHOP780206'].notes\n#\"\"\n\nrecord_correlation_coefficient = aaindex1['CHOP780206']['correlation_coefficient']\nrecord_correlation_coefficient = aaindex1['CHOP780206'].correlation_coefficient\n#{}\n\nrecord_pmid = aaindex1['CHOP780206']['pmid'] \nrecord_pmid = aaindex1['CHOP780206'].pmid\n#364941\n\nrecord_category = aaindex1['CHOP780206']['category']\nrecord_category = aaindex1['CHOP780206'].category\n#sec_struct\n```\n\n### Get total number of AAindex1 records\n```python\n#get total number of records in AAI database\naaindex1.num_records()\n```\n\n### Get list of all AAindex1 record names\n```python\n#get list of all AAindex1 record names\naaindex1.record_names()\n```\n\n## AAIndex2 Usage\n```python\n# from aaindex import aaindex1\nfrom aaindex import aaindex2\n# from aaindex import aaindex3\n```\n## AAIndex3 Usage\n```python\n# from aaindex import aaindex1\n# from aaindex import aaindex2\nfrom aaindex import aaindex3\n```\nDirectories \ud83d\udcc1\n--------------\n* `/tests` - unit and integration tests for `aaindex` package.\n* `/aaindex` - source code and all required external data files for package.\n* `/images` - images used throughout README.\n* `/docs` - `aaindex` documentation.\n\nTests \ud83e\uddea\n--------\nTo run all tests, from the main `aaindex` folder run:\n```\npython3 -m unittest discover tests\n```\n\nContact\n-------\nIf you have any questions or comments, please contact amckenna41@qub.ac.uk or raise an issue on the [Issues][Issues] tab.\n\nLicense\n-------\nDistributed under the MIT License. See `LICENSE` for more details. \n\nReferences\n----------\n\\[1\\]: Shuichi Kawashima, Minoru Kanehisa, AAindex: Amino Acid index database, Nucleic Acids Research, Volume 28, Issue 1, 1 January 2000, Page 374, https://doi.org/10.1093/nar/28.1.374 <br>\n\\[2\\]: https://www.genome.jp/aaindex/ \n\n[Back to top](#TOP)\n\n[python]: https://www.python.org/downloads/release/python-360/\n[aaindex]: https://github.com/amckenna41/aaindex\n[requests]: https://requests.readthedocs.io/en/latest/\n[numpy]: https://numpy.org/\n[PyPi]: https://pypi.org/project/aaindex/\n[demo]: https://colab.research.google.com/drive/1dccV_n1BRMiU8W13F9PPXbSaFzvOdQLC?usp=sharing\n[Issues]: https://github.com/amckenna41/aaindex/issues\n\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "A lightweight Python software package for accessing the data in the various AAIndex databases, which represent the physiochemical, biochemical and structural properties of amino acids as numerical indices.",
"version": "1.1.1",
"project_urls": {
"Download": "https://github.com/amckenna41/aaindex/archive/refs/heads/main.zip",
"Homepage": "https://github.com/amckenna41/aaindex"
},
"split_keywords": [
"amino acid index",
"aaindex",
"bioinformatics",
"protein engineering",
"python",
"pypi",
"physiochemical properties",
"biochemical properties",
"proteins",
"protein structure prediction",
"pysar"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "fa8e4dcaa180bc805257ef47480120500f2df9e684d67eb648f68c19601390ba",
"md5": "cfe86dc40711160d0eccab93cec0a35a",
"sha256": "af7798bb6cfdda4c04c56901e49c27dda9a4d26dd78a38eef3c1113edca2d789"
},
"downloads": -1,
"filename": "aaindex-1.1.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "cfe86dc40711160d0eccab93cec0a35a",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 372883,
"upload_time": "2023-09-18T21:52:34",
"upload_time_iso_8601": "2023-09-18T21:52:34.118211Z",
"url": "https://files.pythonhosted.org/packages/fa/8e/4dcaa180bc805257ef47480120500f2df9e684d67eb648f68c19601390ba/aaindex-1.1.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2023-09-18 21:52:34",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "amckenna41",
"github_project": "aaindex",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"circle": true,
"lcname": "aaindex"
}
