# abcoder
Agentic backend coder - A Jupyter notebook manager with MCP (Model Context Protocol) integration for AI-assisted code execution and bioinformatics workflows.
**Supports multiple Jupyter kernels (Python, R, etc.) for parallel notebook management.**
## 🪩 What can it do?
- **Jupyter Notebook Management**: Create, switch between, and manage multiple Jupyter notebooks
- **Multi-kernel Support**: Manage and run code in multiple Jupyter kernels (e.g., Python, R) simultaneously
- **Code Execution**: Execute single-step or multi-step code in Jupyter kernels
- **Variable Backup**: Safely backup variables before code execution to prevent data loss
- **API Documentation**: Query function and API documentation directly from the kernel
- **Output Handling**: Capture and display execution results, errors, and generated figures
- **Bioinformatics Integration**: Designed for bioinformatics workflows with support for common libraries like scanpy, pandas, numpy, etc.
## ❓ Who is this for?
- **Bioinformaticians** who want AI assistance in their Jupyter workflows
- **AI developers** building agents that need to execute code in Jupyter environments
- **Researchers** who want to integrate AI tools with their computational notebooks
- **Anyone** who wants to use natural language to control Jupyter notebook execution
## 🌐 Where to use it?
You can use abcoder in most AI clients, plugins, or agent frameworks that support the MCP:
- **AI clients**: Cherry Studio, Claude Desktop, etc.
- **Plugins**: Cline, etc.
- **Agent frameworks**: Agno, etc.
## 🎬 Demo
A demo showing AI-assisted bioinformatics analysis in a Jupyter notebook using natural language commands through abcoder.
[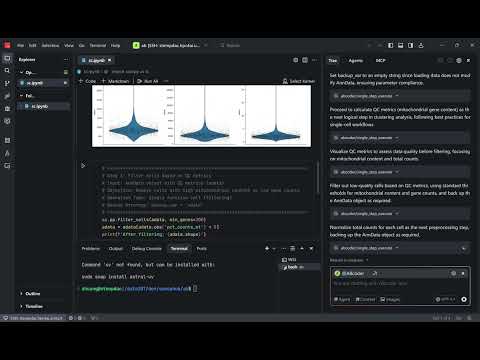](https://youtu.be/3jtXIeapslI)
**Click the image above to watch the demo video**
## 📚 Documentation
For complete documentation, visit: https://github.com/huang-sh/abcoder
## 🏎️ Quickstart
### Install
Install from PyPI:
```bash
pip install abcoder
```
Test the installation:
```bash
abcoder run
```
### Configuration
#### Run abcoder locally
First, check the installation path:
```bash
which abcoder
# Example output: /home/user/bin/abcoder
```
Configure your MCP client:
```json
{
"mcpServers": {
"abcoder": {
"command": "/home/user/bin/abcoder",
"args": ["run"]
}
}
}
```
#### Run abcoder remotely
Start the server on your remote machine:
```bash
abcoder run --transport shttp --port 8000
```
Configure your local MCP client:
```json
{
"mcpServers": {
"abcoder": {
"url": "http://localhost:8000/mcp"
}
}
}
```
## 🛠️ Available Tools
### Notebook Management
- `create_notebook`: Create a new Jupyter notebook with specified ID and path
- `switch_active_notebook`: Switch between different notebooks
### Code Execution
- `single_step_execute`: Execute a single code block
- `multi_step_execute`: Execute multiple code steps with cell addition
- `query_api_doc`: Query function documentation from the kernel
### Features
- **Variable Backup**: Automatically backup variables before execution
- **Error Handling**: Comprehensive error capture and reporting
- **Output Display**: Support for text, images, and other display data
- **Kernel Management**: Automatic kernel lifecycle management
## 🤝 Contributing
If you have any questions, welcome to submit an issue, or contact me(hsh-me@outlook.com). Contributions to the code are also welcome!
Raw data
{
"_id": null,
"home_page": null,
"name": "abcoder",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.10",
"maintainer_email": null,
"keywords": "AI, agent, bioinformatics, llm, mcp, model context protocol, scRNA-seq, single cell",
"author": null,
"author_email": "shuang <hsh-me@outlook.com>",
"download_url": "https://files.pythonhosted.org/packages/f5/99/6162a70b1e5e532f73964a324348710ca565282ec2479700d8b7c359e214/abcoder-0.3.1.tar.gz",
"platform": null,
"description": "# abcoder\n\nAgentic backend coder - A Jupyter notebook manager with MCP (Model Context Protocol) integration for AI-assisted code execution and bioinformatics workflows.\n\n**Supports multiple Jupyter kernels (Python, R, etc.) for parallel notebook management.**\n\n## \ud83e\udea9 What can it do?\n\n- **Jupyter Notebook Management**: Create, switch between, and manage multiple Jupyter notebooks\n- **Multi-kernel Support**: Manage and run code in multiple Jupyter kernels (e.g., Python, R) simultaneously\n- **Code Execution**: Execute single-step or multi-step code in Jupyter kernels\n- **Variable Backup**: Safely backup variables before code execution to prevent data loss\n- **API Documentation**: Query function and API documentation directly from the kernel\n- **Output Handling**: Capture and display execution results, errors, and generated figures\n- **Bioinformatics Integration**: Designed for bioinformatics workflows with support for common libraries like scanpy, pandas, numpy, etc.\n\n## \u2753 Who is this for?\n\n- **Bioinformaticians** who want AI assistance in their Jupyter workflows\n- **AI developers** building agents that need to execute code in Jupyter environments\n- **Researchers** who want to integrate AI tools with their computational notebooks\n- **Anyone** who wants to use natural language to control Jupyter notebook execution\n\n## \ud83c\udf10 Where to use it?\n\nYou can use abcoder in most AI clients, plugins, or agent frameworks that support the MCP:\n\n- **AI clients**: Cherry Studio, Claude Desktop, etc.\n- **Plugins**: Cline, etc.\n- **Agent frameworks**: Agno, etc.\n\n## \ud83c\udfac Demo\n\nA demo showing AI-assisted bioinformatics analysis in a Jupyter notebook using natural language commands through abcoder.\n\n[](https://youtu.be/3jtXIeapslI)\n\n**Click the image above to watch the demo video**\n\n## \ud83d\udcda Documentation\n\nFor complete documentation, visit: https://github.com/huang-sh/abcoder\n\n## \ud83c\udfce\ufe0f Quickstart\n\n### Install\n\nInstall from PyPI:\n```bash\npip install abcoder\n```\n\nTest the installation:\n```bash\nabcoder run\n```\n\n### Configuration\n\n#### Run abcoder locally\n\nFirst, check the installation path:\n```bash\nwhich abcoder\n# Example output: /home/user/bin/abcoder\n```\n\nConfigure your MCP client:\n```json\n{\n \"mcpServers\": {\n \"abcoder\": {\n \"command\": \"/home/user/bin/abcoder\",\n \"args\": [\"run\"]\n }\n }\n}\n```\n\n#### Run abcoder remotely\n\nStart the server on your remote machine:\n```bash\nabcoder run --transport shttp --port 8000\n```\n\nConfigure your local MCP client:\n```json\n{\n \"mcpServers\": {\n \"abcoder\": {\n \"url\": \"http://localhost:8000/mcp\"\n }\n }\n}\n```\n\n## \ud83d\udee0\ufe0f Available Tools\n\n### Notebook Management\n- `create_notebook`: Create a new Jupyter notebook with specified ID and path\n- `switch_active_notebook`: Switch between different notebooks\n\n### Code Execution\n- `single_step_execute`: Execute a single code block\n- `multi_step_execute`: Execute multiple code steps with cell addition\n- `query_api_doc`: Query function documentation from the kernel\n\n### Features\n- **Variable Backup**: Automatically backup variables before execution\n- **Error Handling**: Comprehensive error capture and reporting\n- **Output Display**: Support for text, images, and other display data\n- **Kernel Management**: Automatic kernel lifecycle management\n\n\n## \ud83e\udd1d Contributing\n\nIf you have any questions, welcome to submit an issue, or contact me(hsh-me@outlook.com). Contributions to the code are also welcome!",
"bugtrack_url": null,
"license": null,
"summary": "Agentic bioinformatics coder",
"version": "0.3.1",
"project_urls": {
"Documentation": "https://github.com/huang-sh/abcoder",
"Homepage": "https://github.com/huang-sh/abcoder",
"Repository": "https://github.com/huang-sh/abcoder"
},
"split_keywords": [
"ai",
" agent",
" bioinformatics",
" llm",
" mcp",
" model context protocol",
" scrna-seq",
" single cell"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "aac17c658f02e6be1394ffeb0a17d40d0d94bf086bfe793fce395f49fea9059d",
"md5": "d2c8c77cbd82bb89d282810c621eb295",
"sha256": "974bf9b48fc4fc25403e753b11fb8fd3aacf2b6b3af656ae1a6266a53dbf7501"
},
"downloads": -1,
"filename": "abcoder-0.3.1-py3-none-any.whl",
"has_sig": false,
"md5_digest": "d2c8c77cbd82bb89d282810c621eb295",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.10",
"size": 17222,
"upload_time": "2025-09-05T09:38:27",
"upload_time_iso_8601": "2025-09-05T09:38:27.153617Z",
"url": "https://files.pythonhosted.org/packages/aa/c1/7c658f02e6be1394ffeb0a17d40d0d94bf086bfe793fce395f49fea9059d/abcoder-0.3.1-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "f5996162a70b1e5e532f73964a324348710ca565282ec2479700d8b7c359e214",
"md5": "3d6f18bacbf1f6c4f6e056b3376ad37a",
"sha256": "a109f8f69452ba8b67f4b25cd48928b4d208e4dd7f5fdb2b7684493c9e6f6e8d"
},
"downloads": -1,
"filename": "abcoder-0.3.1.tar.gz",
"has_sig": false,
"md5_digest": "3d6f18bacbf1f6c4f6e056b3376ad37a",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.10",
"size": 178199,
"upload_time": "2025-09-05T09:38:28",
"upload_time_iso_8601": "2025-09-05T09:38:28.596764Z",
"url": "https://files.pythonhosted.org/packages/f5/99/6162a70b1e5e532f73964a324348710ca565282ec2479700d8b7c359e214/abcoder-0.3.1.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-09-05 09:38:28",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "huang-sh",
"github_project": "abcoder",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"lcname": "abcoder"
}
