[](https://pypi.org/project/flowpaths/)
[](https://github.com/algbio/flowpaths/blob/main/LICENSE)
[](https://github.com/algbio/flowpaths/actions/workflows/dx3-tests.yml)
[](https://codecov.io/gh/algbio/flowpaths)
# The _flowpaths_ Python Package
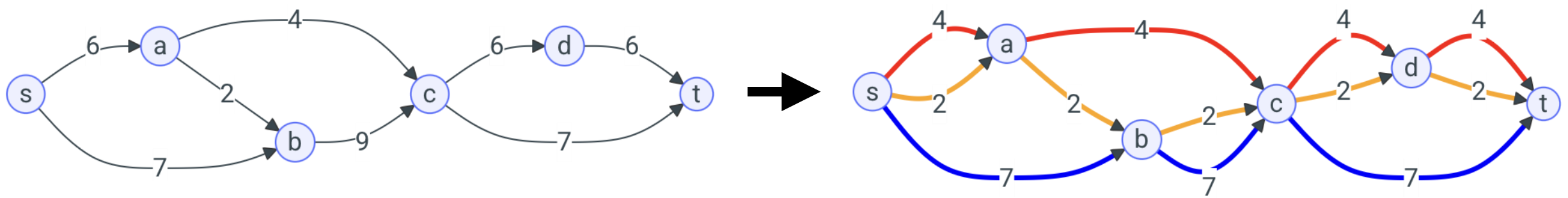
This package implements solvers for decomposing weighted directed graphs into weighted paths or walks, based on (Mixed) Integer Linear Programming ((M)ILP) formulations. It supports both acyclic graphs (DAGs, decomposed into paths) and general graphs with cycles (decomposed into walks), and makes it easy to create new decomposition models.
### Installation
```bash
pip install flowpaths
```
### Documentation
The documentation is available at [algbio.github.io/flowpaths/](https://algbio.github.io/flowpaths/).
### Requirements
- Python >= 3.8
- Dependencies installed automatically: networkx, highspy, graphviz, numpy
- Optional: gurobipy (to use Gurobi instead of the default HiGHS)
### Basic usage example
```python
import flowpaths as fp
import networkx as nx
# Create a simple graph
graph = nx.DiGraph()
graph.add_edge("s", "a", flow=2)
graph.add_edge("a", "t", flow=2)
graph.add_edge("s", "b", flow=5)
graph.add_edge("b", "t", flow=5)
# ...
# Create a Minimum Flow Decomposition solver
mfd_solver = fp.MinFlowDecomp(graph, flow_attr="flow")
mfd_solver.solve() # We solve it
if mfd_solver.is_solved(): # We get the solution
print(mfd_solver.get_solution())
# {'paths': [['s', 'b', 't'], ['s', 'a', 't']], 'weights': [5, 2]}
```
For graphs with cycles, use the cyclic variants which return walks rather than simple paths:
```python
import flowpaths as fp
import networkx as nx
G = nx.DiGraph()
G.add_edge("s", "a", flow=1)
G.add_edge("a", "b", flow=2) # part of a cycle
G.add_edge("b", "a", flow=2) # part of a cycle
G.add_edge("a", "t", flow=1)
mfd_solver = fp.MinFlowDecompCycles(G, flow_attr="flow")
mfd_solver.solve()
if mfd_solver.is_solved():
print(mfd_solver.get_solution())
# {'walks': [['s', 'a', 'b', 'a', 'b', 'a', 't']], 'weights': [1]}
```
### Design principles
1. **Easy to use**: You pass a directed graph (as a [networkx](https://networkx.org) [DiGraph](https://networkx.org/documentation/stable/reference/classes/digraph.html)), and the solvers return optimal weighted paths (or walks for cyclic models). See the [examples/](https://github.com/algbio/flowpaths/tree/main/examples) folder.
2. **It just works**: You do not need to install an (M)ILP solver. This is possible thanks to the fast open source solver [HiGHS](https://highs.dev), which gets installed once you install this package.
- If you have a [Gurobi](https://www.gurobi.com/solutions/gurobi-optimizer/) license ([free for academic users](https://www.gurobi.com/features/academic-named-user-license/)), you can install the [gurobipy Python package](https://support.gurobi.com/hc/en-us/articles/360044290292-How-do-I-install-Gurobi-for-Python), and then you can run the Gurobi solver instead of the default HiGHS solver by just passing the entry `"external_solver": "gurobi"` in the `solver_options` dictionary.
3. **Easy to implement other decomposition models**:
- For DAGs, use the abstract class `AbstractPathModelDAG`, which encodes a given number of paths. See docs: [Abstract Path Model](https://algbio.github.io/flowpaths/abstract-path-model.html).
- For general directed graphs with cycles, use `AbstractWalkModelDiGraph`, which encodes a given number of walks. See docs: [Abstract Walk Model](https://algbio.github.io/flowpaths/abstract-walk-model.html).
You can inherit from these classes to add weights and model-specific constraints/objectives. See [a basic example](examples/inexact_flow_solver.py). These abstract classes interface with a wrapper for popular MILP solvers, so you don't need to worry about solver-specific details.
4. **Fast**: Having solvers implemented using `AbstractPathModelDAG` or `AbstractWalkModelDiGraph` means that any optimization to the path-/walk-finding mechanisms benefits **all** solvers that inherit from these classes. We implement some "safety optimizations" described in [this paper](https://doi.org/10.48550/arXiv.2411.03871), based on ideas first introduced in [this paper](https://doi.org/10.4230/LIPIcs.SEA.2024.14), which can provide up to **1000x speedups**, depending on the graph instance, while preserving global optimality (under some simple assumptions).
5. **Flexible inputs**: The models support graphs with flows/weights on either edges or nodes, and additional real use-case input features, such as [subpathconstraints](https://algbio.github.io/flowpaths/subpath-constraints.html) or [subset constraints](https://algbio.github.io/flowpaths/subset-constraints.html).
### Models currently implemented
- [**Minimum Flow Decomposition**](https://algbio.github.io/flowpaths/minimum-flow-decomposition.html): Given a graph with flow values on its edges (i.e. at every node different from source or sink the flow entering the node is equal to the flow exiting the node), find the minimum number of weighted paths / walks such that, for every edge, the sum of the weights of the paths going through the edge equals the flow value of the edge.
- [**_k_-Least Absolute Errors**](https://algbio.github.io/flowpaths/k-least-absolute-errors.html): Given a graph with weights on its edges, and a number $k$, find $k$ weighted paths / walks such that the sum of the absolute errors of each edge is minimized.
- The *error of an edge* is defined as the weight of the edge minus the sum of the weights of the paths / walks going through it.
- [**_k_-Minimum Path Error**](https://algbio.github.io/flowpaths/k-min-path-error.html): Given a graph with weights on its edges, and a number $k$, find $k$ weighted paths / walks, with associated *slack* values, such that:
- The error of each edge (defined as in $k$-Least Absolute Errors above) is at most the sum of the slacks of the paths / walks going through the edge, and
- The sum of path / walk slacks is minimized.
- [**Minimum Path Cover**](https://algbio.github.io/flowpaths/minimum-path-cover.html): Given a graph and node sets _S_ and _T_, find a minimum number of _S-T_ paths (if the graph is acyclic) or _S-T_ walks (if the graph has cycles) such that every edge appears in in at least one path or walk.
### Contributing
Contributions are welcome! Please read the [CONTRIBUTING.md](https://github.com/algbio/flowpaths/blob/main/CONTRIBUTING.md) guide for how to set up a dev environment, run tests locally, and build/preview the documentation with [Material for MkDocs](https://squidfunk.github.io/mkdocs-material/).
### License and support
- License: MIT
- Issues: [https://github.com/algbio/flowpaths/issues](https://github.com/algbio/flowpaths/issues)
Raw data
{
"_id": null,
"home_page": null,
"name": "flowpaths",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.8",
"maintainer_email": null,
"keywords": null,
"author": "Graph Algorithms and Bioinformatics Group @ University of Helsinki, and external collaborators",
"author_email": null,
"download_url": "https://files.pythonhosted.org/packages/43/23/28df8e6e53fec73e28cb0ca866e3e66951ce054c30eeb0c10d4c65e42819/flowpaths-0.2.6.tar.gz",
"platform": null,
"description": "[](https://pypi.org/project/flowpaths/)\n[](https://github.com/algbio/flowpaths/blob/main/LICENSE)\n[](https://github.com/algbio/flowpaths/actions/workflows/dx3-tests.yml)\n[](https://codecov.io/gh/algbio/flowpaths)\n\n# The _flowpaths_ Python Package\n\nThis package implements solvers for decomposing weighted directed graphs into weighted paths or walks, based on (Mixed) Integer Linear Programming ((M)ILP) formulations. It supports both acyclic graphs (DAGs, decomposed into paths) and general graphs with cycles (decomposed into walks), and makes it easy to create new decomposition models.\n\n\n\n### Installation\n\n```bash\npip install flowpaths\n```\n\n### Documentation\n\nThe documentation is available at [algbio.github.io/flowpaths/](https://algbio.github.io/flowpaths/).\n\n### Requirements\n\n- Python >= 3.8\n- Dependencies installed automatically: networkx, highspy, graphviz, numpy\n- Optional: gurobipy (to use Gurobi instead of the default HiGHS)\n\n### Basic usage example\n\n```python\nimport flowpaths as fp\nimport networkx as nx\n\n# Create a simple graph\ngraph = nx.DiGraph()\ngraph.add_edge(\"s\", \"a\", flow=2)\ngraph.add_edge(\"a\", \"t\", flow=2)\ngraph.add_edge(\"s\", \"b\", flow=5)\ngraph.add_edge(\"b\", \"t\", flow=5)\n# ...\n\n# Create a Minimum Flow Decomposition solver\nmfd_solver = fp.MinFlowDecomp(graph, flow_attr=\"flow\") \n\nmfd_solver.solve() # We solve it\n\nif mfd_solver.is_solved(): # We get the solution\n print(mfd_solver.get_solution())\n # {'paths': [['s', 'b', 't'], ['s', 'a', 't']], 'weights': [5, 2]}\n```\n\nFor graphs with cycles, use the cyclic variants which return walks rather than simple paths:\n\n```python\nimport flowpaths as fp\nimport networkx as nx\n\nG = nx.DiGraph()\nG.add_edge(\"s\", \"a\", flow=1)\nG.add_edge(\"a\", \"b\", flow=2) # part of a cycle\nG.add_edge(\"b\", \"a\", flow=2) # part of a cycle\nG.add_edge(\"a\", \"t\", flow=1)\n\nmfd_solver = fp.MinFlowDecompCycles(G, flow_attr=\"flow\")\nmfd_solver.solve()\nif mfd_solver.is_solved():\n print(mfd_solver.get_solution())\n # {'walks': [['s', 'a', 'b', 'a', 'b', 'a', 't']], 'weights': [1]}\n```\n\n### Design principles\n\n1. **Easy to use**: You pass a directed graph (as a [networkx](https://networkx.org) [DiGraph](https://networkx.org/documentation/stable/reference/classes/digraph.html)), and the solvers return optimal weighted paths (or walks for cyclic models). See the [examples/](https://github.com/algbio/flowpaths/tree/main/examples) folder.\n \n2. **It just works**: You do not need to install an (M)ILP solver. This is possible thanks to the fast open source solver [HiGHS](https://highs.dev), which gets installed once you install this package. \n - If you have a [Gurobi](https://www.gurobi.com/solutions/gurobi-optimizer/) license ([free for academic users](https://www.gurobi.com/features/academic-named-user-license/)), you can install the [gurobipy Python package](https://support.gurobi.com/hc/en-us/articles/360044290292-How-do-I-install-Gurobi-for-Python), and then you can run the Gurobi solver instead of the default HiGHS solver by just passing the entry `\"external_solver\": \"gurobi\"` in the `solver_options` dictionary.\n\n3. **Easy to implement other decomposition models**: \n - For DAGs, use the abstract class `AbstractPathModelDAG`, which encodes a given number of paths. See docs: [Abstract Path Model](https://algbio.github.io/flowpaths/abstract-path-model.html).\n - For general directed graphs with cycles, use `AbstractWalkModelDiGraph`, which encodes a given number of walks. See docs: [Abstract Walk Model](https://algbio.github.io/flowpaths/abstract-walk-model.html).\n \n You can inherit from these classes to add weights and model-specific constraints/objectives. See [a basic example](examples/inexact_flow_solver.py). These abstract classes interface with a wrapper for popular MILP solvers, so you don't need to worry about solver-specific details.\n\n4. **Fast**: Having solvers implemented using `AbstractPathModelDAG` or `AbstractWalkModelDiGraph` means that any optimization to the path-/walk-finding mechanisms benefits **all** solvers that inherit from these classes. We implement some \"safety optimizations\" described in [this paper](https://doi.org/10.48550/arXiv.2411.03871), based on ideas first introduced in [this paper](https://doi.org/10.4230/LIPIcs.SEA.2024.14), which can provide up to **1000x speedups**, depending on the graph instance, while preserving global optimality (under some simple assumptions).\n\n5. **Flexible inputs**: The models support graphs with flows/weights on either edges or nodes, and additional real use-case input features, such as [subpathconstraints](https://algbio.github.io/flowpaths/subpath-constraints.html) or [subset constraints](https://algbio.github.io/flowpaths/subset-constraints.html).\n\n### Models currently implemented\n- [**Minimum Flow Decomposition**](https://algbio.github.io/flowpaths/minimum-flow-decomposition.html): Given a graph with flow values on its edges (i.e. at every node different from source or sink the flow entering the node is equal to the flow exiting the node), find the minimum number of weighted paths / walks such that, for every edge, the sum of the weights of the paths going through the edge equals the flow value of the edge.\n \n- [**_k_-Least Absolute Errors**](https://algbio.github.io/flowpaths/k-least-absolute-errors.html): Given a graph with weights on its edges, and a number $k$, find $k$ weighted paths / walks such that the sum of the absolute errors of each edge is minimized. \n - The *error of an edge* is defined as the weight of the edge minus the sum of the weights of the paths / walks going through it.\n \n- [**_k_-Minimum Path Error**](https://algbio.github.io/flowpaths/k-min-path-error.html): Given a graph with weights on its edges, and a number $k$, find $k$ weighted paths / walks, with associated *slack* values, such that:\n - The error of each edge (defined as in $k$-Least Absolute Errors above) is at most the sum of the slacks of the paths / walks going through the edge, and\n - The sum of path / walk slacks is minimized.\n \n- [**Minimum Path Cover**](https://algbio.github.io/flowpaths/minimum-path-cover.html): Given a graph and node sets _S_ and _T_, find a minimum number of _S-T_ paths (if the graph is acyclic) or _S-T_ walks (if the graph has cycles) such that every edge appears in in at least one path or walk.\n\n### Contributing\n\nContributions are welcome! Please read the [CONTRIBUTING.md](https://github.com/algbio/flowpaths/blob/main/CONTRIBUTING.md) guide for how to set up a dev environment, run tests locally, and build/preview the documentation with [Material for MkDocs](https://squidfunk.github.io/mkdocs-material/).\n\n### License and support\n\n- License: MIT\n- Issues: [https://github.com/algbio/flowpaths/issues](https://github.com/algbio/flowpaths/issues)\n",
"bugtrack_url": null,
"license": null,
"summary": "A Python package to quickly decompose weighted graphs into weights paths, under various models.",
"version": "0.2.6",
"project_urls": {
"Documentation": "https://algbio.github.io/flowpaths/",
"Homepage": "https://github.com/algbio/flowpaths",
"Issues": "https://github.com/algbio/flowpaths/issues"
},
"split_keywords": [],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "29d2a7fdd72190f6868287dec4934fa4ff1004443bfb741d35476126ba6890c5",
"md5": "38f209fdd72a529931ec120abe0321ab",
"sha256": "8f3bdafdb356fbd220cb39c038786ca66e7efd6ba422c139a7a81d5aa9e07be2"
},
"downloads": -1,
"filename": "flowpaths-0.2.6-py3-none-any.whl",
"has_sig": false,
"md5_digest": "38f209fdd72a529931ec120abe0321ab",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.8",
"size": 140658,
"upload_time": "2025-09-08T10:55:51",
"upload_time_iso_8601": "2025-09-08T10:55:51.554544Z",
"url": "https://files.pythonhosted.org/packages/29/d2/a7fdd72190f6868287dec4934fa4ff1004443bfb741d35476126ba6890c5/flowpaths-0.2.6-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "432328df8e6e53fec73e28cb0ca866e3e66951ce054c30eeb0c10d4c65e42819",
"md5": "8febf928bb1cb68ef79678e373f4d76d",
"sha256": "bdd1e9ed29afb8acd5a0bbadd0520d194addf52ab7f00851b3eabea6935f6dff"
},
"downloads": -1,
"filename": "flowpaths-0.2.6.tar.gz",
"has_sig": false,
"md5_digest": "8febf928bb1cb68ef79678e373f4d76d",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.8",
"size": 111208,
"upload_time": "2025-09-08T10:55:53",
"upload_time_iso_8601": "2025-09-08T10:55:53.283584Z",
"url": "https://files.pythonhosted.org/packages/43/23/28df8e6e53fec73e28cb0ca866e3e66951ce054c30eeb0c10d4c65e42819/flowpaths-0.2.6.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-09-08 10:55:53",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "algbio",
"github_project": "flowpaths",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [
{
"name": "networkx",
"specs": [
[
">=",
"3.4.2"
]
]
},
{
"name": "highspy",
"specs": [
[
">=",
"1.10.0"
]
]
},
{
"name": "graphviz",
"specs": [
[
">=",
"0.20.3"
]
]
},
{
"name": "numpy",
"specs": [
[
">=",
"2.2.5"
]
]
}
],
"lcname": "flowpaths"
}
