# Gene Scoring Package
This is a Python package implementing a novel gene set scoring methodology for single-cell RNA sequencing data analysis. It builds upon and extends techniques from widely used single-cell analysis toolkits such as Seurat and Scanpy, introducing several key improvements for more accurate gene set scoring.
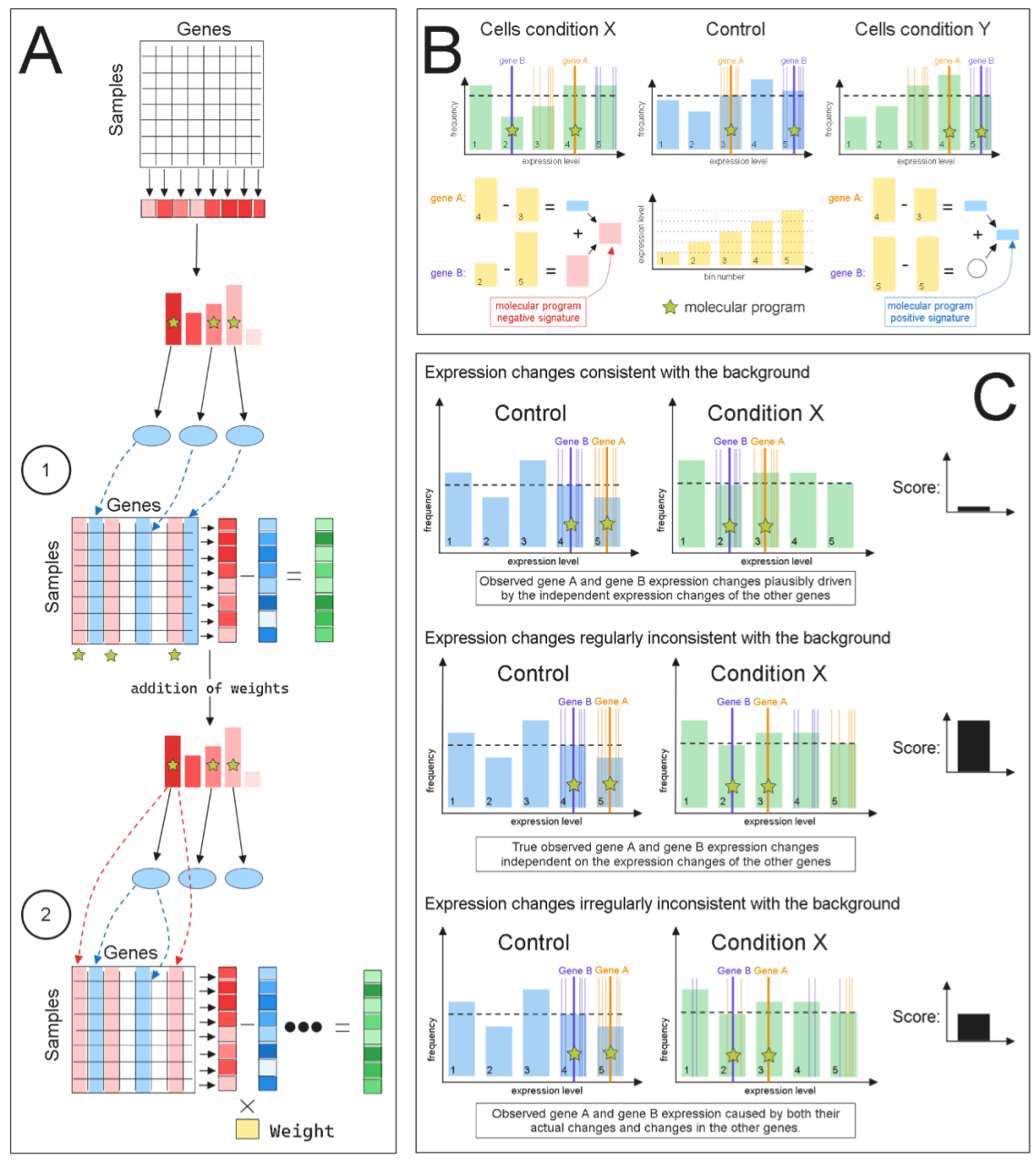
## Key Features
1. **Weighted Gene Contributions**
- Each gene in the evaluated module is assigned an appropriate weight based on its correlation with the phenotype of interest
- Weights can be normalized to sum to 1 (optional)
2. **Individualized Background Selection**
- Background genes are selected independently for each gene of interest
- Control features are selected from the same expression bin in the histogram
- Background can be based on control sample, entire dataset, or a selected pool of genes
3. **Variance-scaled Gene Contribution**
- Individual gene contributions are scaled by their variance under examined conditions
- Helps account for gene expression variability in different conditions
4. **Optimized Computation**
- GPU-accelerated score calculation for improved performance
- Support for sparse matrices and chunked processing for large datasets
- Efficient background gene selection algorithm
## Installation
### Basic Installation
```bash
pip install gene_scoring
```
### Installation with GPU Support
```bash
pip install gene_scoring[gpu]
```
## Mathematical Foundation
The scoring methodology is defined by the following formula:
```
S_c = Σ (w_l / (σ_c,l + ε)) * (X_c,l - X̄_l,control)
```
Where:
- n: number of genes in the gene list
- w_l: weight for gene l
- X_c,l: mean expression value of gene l in condition c
- X̄_l,control: average expression of control genes in the background
- σ_c,l: standard deviation of gene expression in condition c
- ε: small constant to avoid division by zero
## Usage
```python
from gene_scoring import GeneScorer
# Initialize the scorer
scorer = GeneScorer(
normalize_weights=True, # Normalize weights to sum to 1
abs_diff=False, # Use signed difference (not absolute)
weighted=True # Use weighted scoring
)
# Calculate scores
scores = scorer.calculate_scores(
expression_matrix, # Gene expression matrix (genes x cells)
gene_list, # List of genes to score
weights=gene_weights # Optional weights for each gene
)
```
## Parameters
The main parameters for score calculation include:
- `normalize_weights`: If True, weights are normalized to sum to 1
- `abs_diff`: If True, use absolute difference between expression and background
- `weighted`: If True, use weighted scoring (otherwise all weights = 1)
- `ctrl_size`: Number of control genes to use per target gene
- `gpu`: Whether to use GPU acceleration
- `chunk_size`: Size of chunks for processing large datasets
- `random_state`: Random seed for reproducibility
## Requirements
- Python ≥ 3.7
- NumPy ≥ 1.19.0
- Pandas ≥ 1.0.0
- SciPy ≥ 1.5.0
- CuPy ≥ 9.0.0 (optional, for GPU support)
## License
This project is licensed under the MIT License - see the LICENSE file for details.
Raw data
{
"_id": null,
"home_page": "https://github.com/michal7kw/GeneScore",
"name": "gene-scoring",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.7",
"maintainer_email": null,
"keywords": "bioinformatics, single-cell, RNA-seq, gene scoring, GPU acceleration",
"author": "Michal Kubacki",
"author_email": "michal.kubacki@example.com",
"download_url": "https://files.pythonhosted.org/packages/b2/b9/928daa66fe02d27f3549c102207142f0858313077eb9383df99acd503838/gene_scoring-0.1.4.tar.gz",
"platform": null,
"description": "# Gene Scoring Package\n\nThis is a Python package implementing a novel gene set scoring methodology for single-cell RNA sequencing data analysis. It builds upon and extends techniques from widely used single-cell analysis toolkits such as Seurat and Scanpy, introducing several key improvements for more accurate gene set scoring.\n\n\n\n## Key Features\n\n1. **Weighted Gene Contributions**\n - Each gene in the evaluated module is assigned an appropriate weight based on its correlation with the phenotype of interest\n - Weights can be normalized to sum to 1 (optional)\n\n2. **Individualized Background Selection**\n - Background genes are selected independently for each gene of interest\n - Control features are selected from the same expression bin in the histogram\n - Background can be based on control sample, entire dataset, or a selected pool of genes\n\n3. **Variance-scaled Gene Contribution**\n - Individual gene contributions are scaled by their variance under examined conditions\n - Helps account for gene expression variability in different conditions\n\n4. **Optimized Computation**\n - GPU-accelerated score calculation for improved performance\n - Support for sparse matrices and chunked processing for large datasets\n - Efficient background gene selection algorithm\n\n## Installation\n\n### Basic Installation\n\n```bash\npip install gene_scoring\n```\n\n### Installation with GPU Support\n\n```bash\npip install gene_scoring[gpu]\n```\n\n## Mathematical Foundation\n\nThe scoring methodology is defined by the following formula:\n\n```\nS_c = \u03a3 (w_l / (\u03c3_c,l + \u03b5)) * (X_c,l - X\u0304_l,control)\n```\n\nWhere:\n- n: number of genes in the gene list\n- w_l: weight for gene l\n- X_c,l: mean expression value of gene l in condition c\n- X\u0304_l,control: average expression of control genes in the background\n- \u03c3_c,l: standard deviation of gene expression in condition c\n- \u03b5: small constant to avoid division by zero\n\n## Usage\n\n```python\nfrom gene_scoring import GeneScorer\n\n# Initialize the scorer\nscorer = GeneScorer(\n normalize_weights=True, # Normalize weights to sum to 1\n abs_diff=False, # Use signed difference (not absolute)\n weighted=True # Use weighted scoring\n)\n\n# Calculate scores\nscores = scorer.calculate_scores(\n expression_matrix, # Gene expression matrix (genes x cells)\n gene_list, # List of genes to score\n weights=gene_weights # Optional weights for each gene\n)\n```\n\n## Parameters\n\nThe main parameters for score calculation include:\n\n- `normalize_weights`: If True, weights are normalized to sum to 1\n- `abs_diff`: If True, use absolute difference between expression and background\n- `weighted`: If True, use weighted scoring (otherwise all weights = 1)\n- `ctrl_size`: Number of control genes to use per target gene\n- `gpu`: Whether to use GPU acceleration\n- `chunk_size`: Size of chunks for processing large datasets\n- `random_state`: Random seed for reproducibility\n\n## Requirements\n\n- Python \u2265 3.7\n- NumPy \u2265 1.19.0\n- Pandas \u2265 1.0.0\n- SciPy \u2265 1.5.0\n- CuPy \u2265 9.0.0 (optional, for GPU support)\n\n## License\n\nThis project is licensed under the MIT License - see the LICENSE file for details.\n",
"bugtrack_url": null,
"license": null,
"summary": "Gene set scoring package for single-cell RNA sequencing data with GPU acceleration.",
"version": "0.1.4",
"project_urls": {
"Bug Reports": "https://github.com/michal7kw/GeneScore/issues",
"Documentation": "https://github.com/michal7kw/GeneScore#readme",
"Homepage": "https://github.com/michal7kw/GeneScore",
"Source Code": "https://github.com/michal7kw/GeneScore"
},
"split_keywords": [
"bioinformatics",
" single-cell",
" rna-seq",
" gene scoring",
" gpu acceleration"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "ae69eff58edf92e122dbdd8ea74546aeab12c16c529467f9f1c016486196e017",
"md5": "01b1f5be6526b22ff4705644a4044b7f",
"sha256": "e5baba63f78f0062dd6adf3206cc96bf8122307d7961191575fd8ba47591917b"
},
"downloads": -1,
"filename": "gene_scoring-0.1.4-py3-none-any.whl",
"has_sig": false,
"md5_digest": "01b1f5be6526b22ff4705644a4044b7f",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.7",
"size": 2963,
"upload_time": "2025-02-03T07:05:54",
"upload_time_iso_8601": "2025-02-03T07:05:54.826215Z",
"url": "https://files.pythonhosted.org/packages/ae/69/eff58edf92e122dbdd8ea74546aeab12c16c529467f9f1c016486196e017/gene_scoring-0.1.4-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "b2b9928daa66fe02d27f3549c102207142f0858313077eb9383df99acd503838",
"md5": "7e3a12530519970af60d231c0dc7e87c",
"sha256": "c6bfcc1a3574d8a9ec860443dca37d55b4b9bcbcca028e04bc66aa8cc70b3e10"
},
"downloads": -1,
"filename": "gene_scoring-0.1.4.tar.gz",
"has_sig": false,
"md5_digest": "7e3a12530519970af60d231c0dc7e87c",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.7",
"size": 390764,
"upload_time": "2025-02-03T07:05:57",
"upload_time_iso_8601": "2025-02-03T07:05:57.382930Z",
"url": "https://files.pythonhosted.org/packages/b2/b9/928daa66fe02d27f3549c102207142f0858313077eb9383df99acd503838/gene_scoring-0.1.4.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-02-03 07:05:57",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "michal7kw",
"github_project": "GeneScore",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"lcname": "gene-scoring"
}
