# gget
[](https://pypi.org/project/gget)
[](https://pepy.tech/project/gget)
[](https://anaconda.org/bioconda/gget)
[](LICENSE)



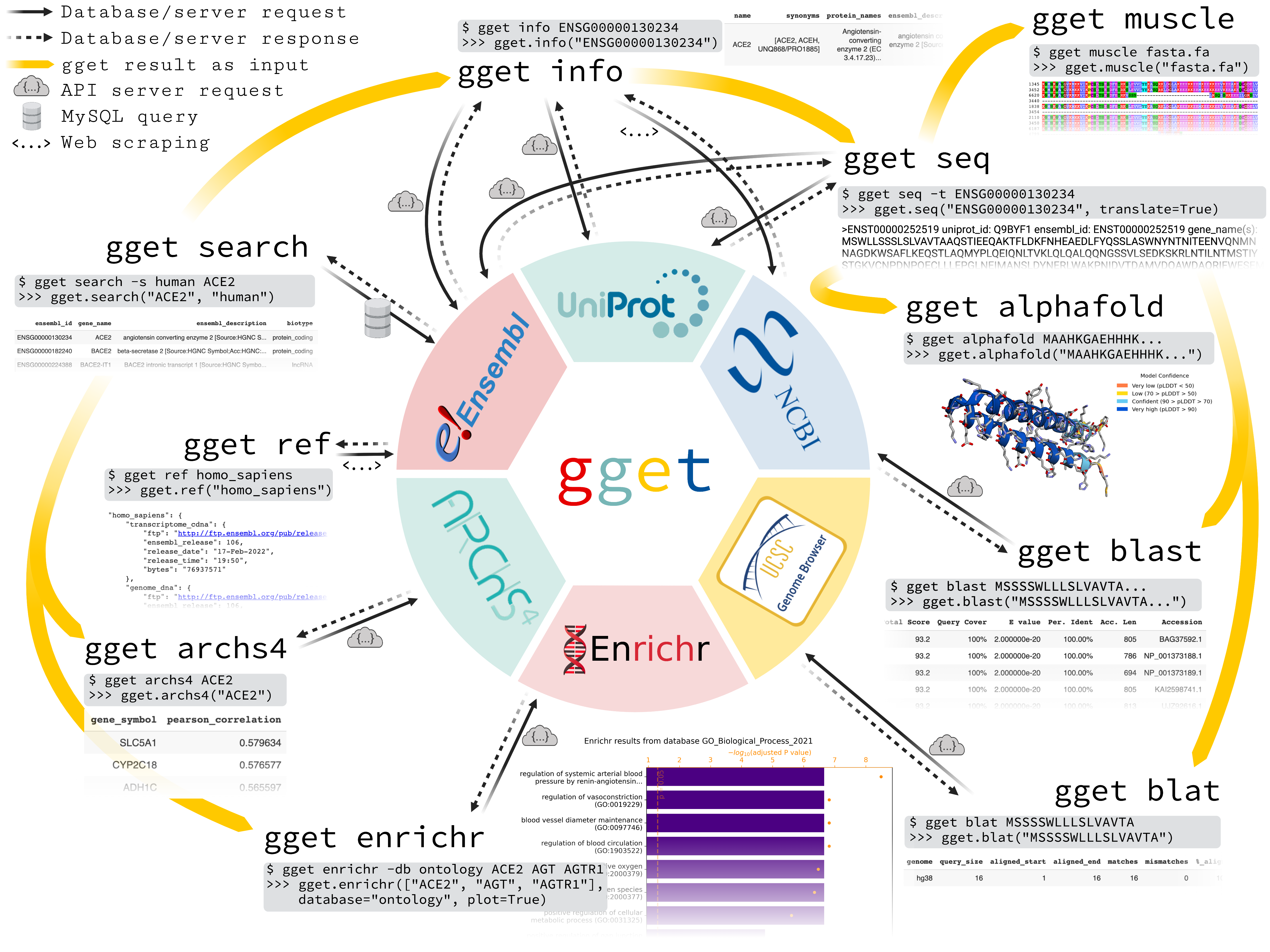
`gget` is a free, open-source command-line tool and Python package that enables efficient querying of genomic databases. `gget` consists of a collection of separate but interoperable modules, each designed to facilitate one type of database querying in a single line of code.
If you use `gget` in a publication, please [cite*](https://pachterlab.github.io/gget/en/cite.html):
```
Luebbert, L., & Pachter, L. (2023). Efficient querying of genomic reference databases with gget. Bioinformatics. https://doi.org/10.1093/bioinformatics/btac836
```
Read the article here: https://doi.org/10.1093/bioinformatics/btac836
# Installation
```bash
uv pip install gget
```
or
```bash
pip install --upgrade gget
```
For use in Jupyter Lab / Google Colab:
```python
# Python
import gget
```
# [🔗 Manual](https://pachterlab.github.io/gget)
# 🪄 Quick start guide
Command line:
```bash
# Fetch all Homo sapiens reference and annotation FTPs from the latest Ensembl release
$ gget ref homo_sapiens
# Get Ensembl IDs of human genes with "ace2" or "angiotensin converting enzyme 2" in their name/description
$ gget search -s homo_sapiens 'ace2' 'angiotensin converting enzyme 2'
# Look up gene ENSG00000130234 (ACE2) and its transcript ENST00000252519
$ gget info ENSG00000130234 ENST00000252519
# Fetch the amino acid sequence of the canonical transcript of gene ENSG00000130234
$ gget seq --translate ENSG00000130234
# Quickly find the genomic location of (the start of) that amino acid sequence
$ gget blat MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
# BLAST (the start of) that amino acid sequence
$ gget blast MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
# Align multiple nucleotide or amino acid sequences against each other (also accepts path to FASTA file)
$ gget muscle MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS
# Align one or more amino acid sequences against a reference (containing one or more sequences) (local BLAST) (also accepts paths to FASTA files)
$ gget diamond MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS -ref MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS
# Use Enrichr for an ontology analysis of a list of genes
$ gget enrichr -db ontology ACE2 AGT AGTR1 ACE AGTRAP AGTR2 ACE3P
# Get the human tissue expression of gene ACE2
$ gget archs4 -w tissue ACE2
# Get the protein structure (in PDB format) of ACE2 as stored in the Protein Data Bank (PDB ID returned by gget info)
$ gget pdb 1R42 -o 1R42.pdb
# Find Eukaryotic Linear Motifs (ELMs) in a protein sequence
$ gget setup elm # setup only needs to be run once
$ gget elm -o results MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
# Fetch a scRNAseq count matrix (AnnData format) based on specified gene(s), tissue(s), and cell type(s) (default species: human)
$ gget setup cellxgene # setup only needs to be run once
$ gget cellxgene --gene ACE2 SLC5A1 --tissue lung --cell_type 'mucus secreting cell' -o example_adata.h5ad
# Predict the protein structure of GFP from its amino acid sequence
$ gget setup alphafold # setup only needs to be run once
$ gget alphafold MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK
```
Python (Jupyter Lab / Google Colab):
```python
import gget
gget.ref("homo_sapiens")
gget.search(["ace2", "angiotensin converting enzyme 2"], "homo_sapiens")
gget.info(["ENSG00000130234", "ENST00000252519"])
gget.seq("ENSG00000130234", translate=True)
gget.blat("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.blast("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.muscle(["MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", "MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS"])
gget.diamond("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", reference="MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS")
gget.enrichr(["ACE2", "AGT", "AGTR1", "ACE", "AGTRAP", "AGTR2", "ACE3P"], database="ontology", plot=True)
gget.archs4("ACE2", which="tissue")
gget.pdb("1R42", save=True)
gget.setup("elm") # setup only needs to be run once
ortho_df, regex_df = gget.elm("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.setup("cellxgene") # setup only needs to be run once
gget.cellxgene(gene = ["ACE2", "SLC5A1"], tissue = "lung", cell_type = "mucus secreting cell")
gget.setup("alphafold") # setup only needs to be run once
gget.alphafold("MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK")
```
Call `gget` from R using [reticulate](https://rstudio.github.io/reticulate/):
```r
system("pip install gget")
install.packages("reticulate")
library(reticulate)
gget <- import("gget")
gget$ref("homo_sapiens")
gget$search(list("ace2", "angiotensin converting enzyme 2"), "homo_sapiens")
gget$info(list("ENSG00000130234", "ENST00000252519"))
gget$seq("ENSG00000130234", translate=TRUE)
gget$blat("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget$blast("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget$muscle(list("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", "MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS"), out="out.afa")
gget$diamond("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", reference="MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS")
gget$enrichr(list("ACE2", "AGT", "AGTR1", "ACE", "AGTRAP", "AGTR2", "ACE3P"), database="ontology")
gget$archs4("ACE2", which="tissue")
gget$pdb("1R42", save=TRUE)
```
#### [More tutorials](https://github.com/pachterlab/gget_examples)
Raw data
{
"_id": null,
"home_page": "https://github.com/pachterlab/gget",
"name": "gget",
"maintainer": "Laura Luebbert",
"docs_url": null,
"requires_python": ">=3.7",
"maintainer_email": "lauralubbert@gmail.com",
"keywords": "gget",
"author": "Laura Luebbert",
"author_email": "lauralubbert@gmail.com",
"download_url": "https://files.pythonhosted.org/packages/cb/0b/e92590418ed27181e6fe7b71c41081c76e74070162e0f85ade2d2a801b99/gget-0.29.3.tar.gz",
"platform": null,
"description": "# gget\n[](https://pypi.org/project/gget)\n[](https://pepy.tech/project/gget)\n[](https://anaconda.org/bioconda/gget)\n[](LICENSE)\n\n\n \n\n`gget` is a free, open-source command-line tool and Python package that enables efficient querying of genomic databases. `gget` consists of a collection of separate but interoperable modules, each designed to facilitate one type of database querying in a single line of code. \n \n\n \nIf you use `gget` in a publication, please [cite*](https://pachterlab.github.io/gget/en/cite.html): \n```\nLuebbert, L., & Pachter, L. (2023). Efficient querying of genomic reference databases with gget. Bioinformatics. https://doi.org/10.1093/bioinformatics/btac836\n```\nRead the article here: https://doi.org/10.1093/bioinformatics/btac836 \n\n# Installation\n```bash\nuv pip install gget\n```\nor\n```bash\npip install --upgrade gget\n```\n\nFor use in Jupyter Lab / Google Colab:\n```python\n# Python\nimport gget\n```\n# [\ud83d\udd17 Manual](https://pachterlab.github.io/gget) \n\n# \ud83e\ude84 Quick start guide\nCommand line:\n```bash\n# Fetch all Homo sapiens reference and annotation FTPs from the latest Ensembl release\n$ gget ref homo_sapiens\n\n# Get Ensembl IDs of human genes with \"ace2\" or \"angiotensin converting enzyme 2\" in their name/description\n$ gget search -s homo_sapiens 'ace2' 'angiotensin converting enzyme 2'\n\n# Look up gene ENSG00000130234 (ACE2) and its transcript ENST00000252519\n$ gget info ENSG00000130234 ENST00000252519\n\n# Fetch the amino acid sequence of the canonical transcript of gene ENSG00000130234\n$ gget seq --translate ENSG00000130234\n\n# Quickly find the genomic location of (the start of) that amino acid sequence\n$ gget blat MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\n\n# BLAST (the start of) that amino acid sequence\n$ gget blast MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\n\n# Align multiple nucleotide or amino acid sequences against each other (also accepts path to FASTA file) \n$ gget muscle MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS\n\n# Align one or more amino acid sequences against a reference (containing one or more sequences) (local BLAST) (also accepts paths to FASTA files) \n$ gget diamond MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS -ref MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS \n\n# Use Enrichr for an ontology analysis of a list of genes\n$ gget enrichr -db ontology ACE2 AGT AGTR1 ACE AGTRAP AGTR2 ACE3P\n\n# Get the human tissue expression of gene ACE2\n$ gget archs4 -w tissue ACE2\n\n# Get the protein structure (in PDB format) of ACE2 as stored in the Protein Data Bank (PDB ID returned by gget info)\n$ gget pdb 1R42 -o 1R42.pdb\n\n# Find Eukaryotic Linear Motifs (ELMs) in a protein sequence\n$ gget setup elm # setup only needs to be run once\n$ gget elm -o results MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\n\n# Fetch a scRNAseq count matrix (AnnData format) based on specified gene(s), tissue(s), and cell type(s) (default species: human)\n$ gget setup cellxgene # setup only needs to be run once\n$ gget cellxgene --gene ACE2 SLC5A1 --tissue lung --cell_type 'mucus secreting cell' -o example_adata.h5ad\n\n# Predict the protein structure of GFP from its amino acid sequence\n$ gget setup alphafold # setup only needs to be run once\n$ gget alphafold MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK\n```\nPython (Jupyter Lab / Google Colab):\n```python \nimport gget\ngget.ref(\"homo_sapiens\")\ngget.search([\"ace2\", \"angiotensin converting enzyme 2\"], \"homo_sapiens\")\ngget.info([\"ENSG00000130234\", \"ENST00000252519\"])\ngget.seq(\"ENSG00000130234\", translate=True)\ngget.blat(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\")\ngget.blast(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\")\ngget.muscle([\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\", \"MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS\"])\ngget.diamond(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\", reference=\"MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS\")\ngget.enrichr([\"ACE2\", \"AGT\", \"AGTR1\", \"ACE\", \"AGTRAP\", \"AGTR2\", \"ACE3P\"], database=\"ontology\", plot=True)\ngget.archs4(\"ACE2\", which=\"tissue\")\ngget.pdb(\"1R42\", save=True)\n\ngget.setup(\"elm\") # setup only needs to be run once\northo_df, regex_df = gget.elm(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\")\n\ngget.setup(\"cellxgene\") # setup only needs to be run once\ngget.cellxgene(gene = [\"ACE2\", \"SLC5A1\"], tissue = \"lung\", cell_type = \"mucus secreting cell\")\n\ngget.setup(\"alphafold\") # setup only needs to be run once\ngget.alphafold(\"MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK\")\n```\nCall `gget` from R using [reticulate](https://rstudio.github.io/reticulate/):\n```r\nsystem(\"pip install gget\")\ninstall.packages(\"reticulate\")\nlibrary(reticulate)\ngget <- import(\"gget\")\n\ngget$ref(\"homo_sapiens\")\ngget$search(list(\"ace2\", \"angiotensin converting enzyme 2\"), \"homo_sapiens\")\ngget$info(list(\"ENSG00000130234\", \"ENST00000252519\"))\ngget$seq(\"ENSG00000130234\", translate=TRUE)\ngget$blat(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\")\ngget$blast(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\")\ngget$muscle(list(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\", \"MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS\"), out=\"out.afa\")\ngget$diamond(\"MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS\", reference=\"MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS\")\ngget$enrichr(list(\"ACE2\", \"AGT\", \"AGTR1\", \"ACE\", \"AGTRAP\", \"AGTR2\", \"ACE3P\"), database=\"ontology\")\ngget$archs4(\"ACE2\", which=\"tissue\")\ngget$pdb(\"1R42\", save=TRUE)\n```\n#### [More tutorials](https://github.com/pachterlab/gget_examples)\n",
"bugtrack_url": null,
"license": "BSD-2",
"summary": "Efficient querying of genomic databases.",
"version": "0.29.3",
"project_urls": {
"Homepage": "https://github.com/pachterlab/gget"
},
"split_keywords": [
"gget"
],
"urls": [
{
"comment_text": null,
"digests": {
"blake2b_256": "e00868b2f358e5ee27bdff0d9922112898bdc9736cee120b08994b71fa1b9bdc",
"md5": "401dfac5642388685314365a25eb1bf4",
"sha256": "aad3f34b161a39fe3ce45eb97c2e5a6220257d4f1be63f3dd299e6ff9cf60614"
},
"downloads": -1,
"filename": "gget-0.29.3-py3-none-any.whl",
"has_sig": false,
"md5_digest": "401dfac5642388685314365a25eb1bf4",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.7",
"size": 43143050,
"upload_time": "2025-09-11T22:33:28",
"upload_time_iso_8601": "2025-09-11T22:33:28.354771Z",
"url": "https://files.pythonhosted.org/packages/e0/08/68b2f358e5ee27bdff0d9922112898bdc9736cee120b08994b71fa1b9bdc/gget-0.29.3-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": null,
"digests": {
"blake2b_256": "cb0be92590418ed27181e6fe7b71c41081c76e74070162e0f85ade2d2a801b99",
"md5": "28eae6ceddfd2154f64fa9c7e5935e0b",
"sha256": "5e26301090d2f29ed4677ab95f87d3e07ad581a1d856767f1dbc14ce1e794003"
},
"downloads": -1,
"filename": "gget-0.29.3.tar.gz",
"has_sig": false,
"md5_digest": "28eae6ceddfd2154f64fa9c7e5935e0b",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.7",
"size": 42875516,
"upload_time": "2025-09-11T22:33:32",
"upload_time_iso_8601": "2025-09-11T22:33:32.319184Z",
"url": "https://files.pythonhosted.org/packages/cb/0b/e92590418ed27181e6fe7b71c41081c76e74070162e0f85ade2d2a801b99/gget-0.29.3.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2025-09-11 22:33:32",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "pachterlab",
"github_project": "gget",
"travis_ci": false,
"coveralls": false,
"github_actions": true,
"requirements": [
{
"name": "numpy",
"specs": [
[
">=",
"1.17.2"
]
]
},
{
"name": "pandas",
"specs": [
[
">=",
"1.0.0"
]
]
},
{
"name": "requests",
"specs": [
[
">=",
"2.22.0"
]
]
},
{
"name": "ipython",
"specs": []
},
{
"name": "matplotlib",
"specs": []
},
{
"name": "mysql-connector-python",
"specs": [
[
">=",
"8.0.32"
]
]
},
{
"name": "beautifulsoup4",
"specs": [
[
">=",
"4.10.0"
]
]
},
{
"name": "ipywidgets",
"specs": []
},
{
"name": "tqdm",
"specs": []
},
{
"name": "lxml",
"specs": []
}
],
"lcname": "gget"
}
