# Higher Order Spectral Analysis Toolkit
This package provides a comprehensive set of tools for higher-order spectral
analysis in Python. It includes functions for estimating bicoherence,
bispectrum, and various orders of cumulants.
## Installation
You can install the HOSA toolkit using pip:
```bash
pip install higher-spectrum
```
## Contents
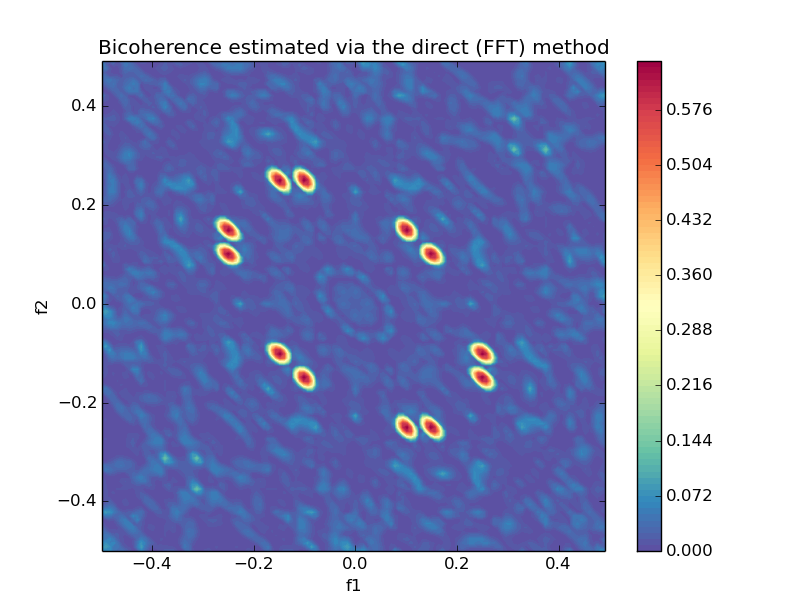
### Bicoherence
```python
from spectrum import bicoherence, plot_bicoherence
bic, waxis = bicoherence(y, nfft=None, window=None, nsamp=None, overlap=None)
plot_bicoherence(bic, waxis)
```
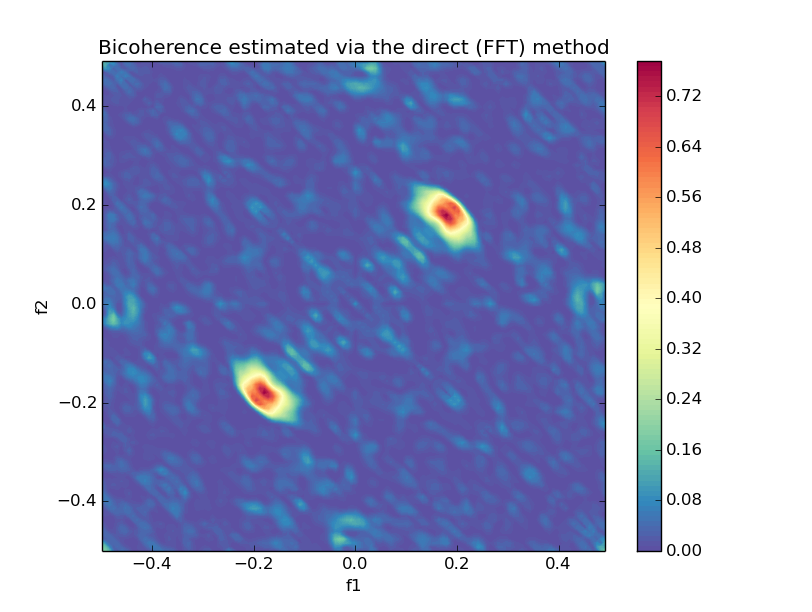
### Cross Bicoherence
```python
from spectrum import bicoherencex, plot_cross_bicoherence
bic, waxis = bicoherencex(w, x, y, nfft=None, window=None, nsamp=None, overlap=None)
plot_cross_bicoherence(bic, waxis)
```
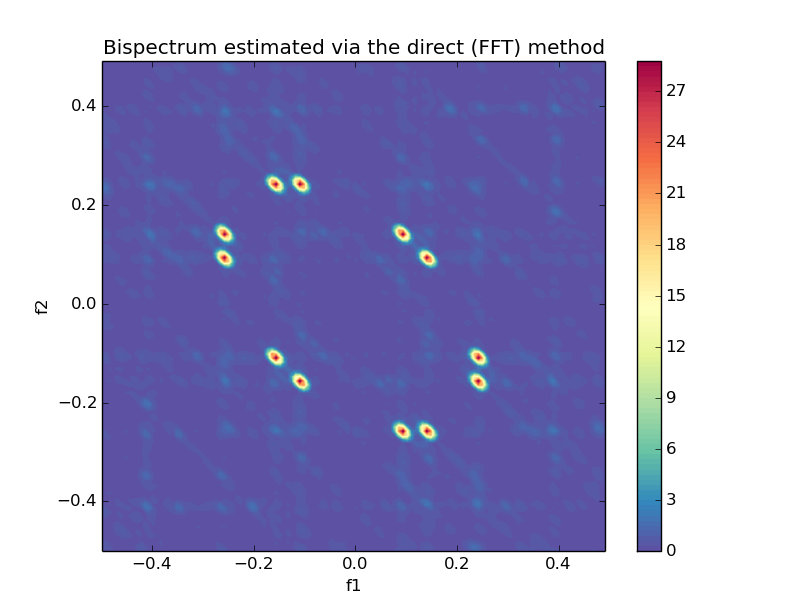
### Bispectrum Direct (using FFT)
```python
from spectrum import bispectrumd, plot_bispectrum
Bspec, waxis = bispectrumd(y, nfft=None, window=None, nsamp=None, overlap=None)
plot_bispectrum(Bspec, waxis)
```
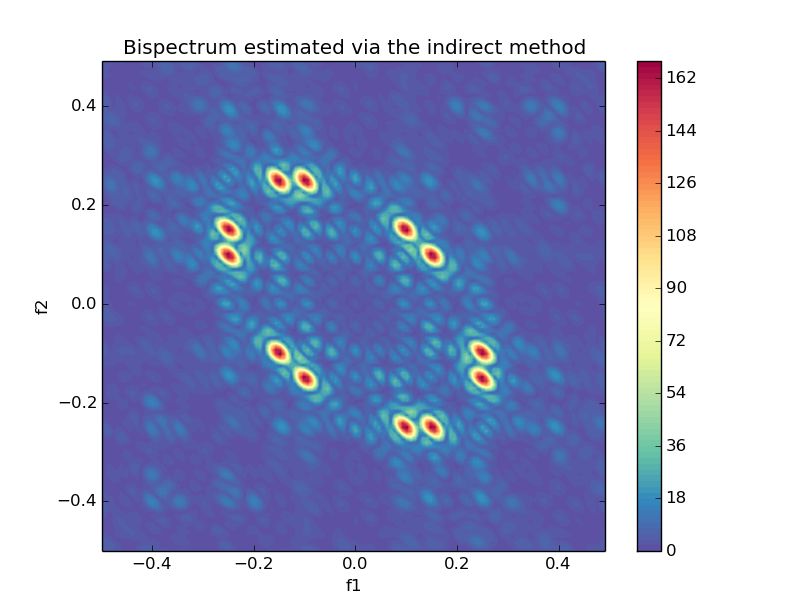
### Bispectrum Indirect
```python
from spectrum import bispectrumi, plot_bispectrum_indirect
Bspec, waxis = bispectrumi(y, nlag=None, nsamp=None, overlap=None, flag='biased', nfft=None, wind='parzen')
plot_bispectrum_indirect(Bspec, waxis)
```
### Cross Bispectrum (Direct)
```python
from spectrum import bispectrumdx, plot_cross_bispectrum
Bspec, waxis = bispectrumdx(x, y, z, nfft=None, window=None, nsamp=None, overlap=None)
plot_cross_bispectrum(Bspec, waxis)
```
### Cumulants (2nd, 3rd, and 4th order)
```python
from spectrum import cumest, plot_cumulant
order = 2 # 2nd order
y_cum = cumest(y, norder=order, maxlag=20, nsamp=None, overlap=0, flag='biased', k1=0, k2=0)
plot_cumulant(np.arange(-20, 21), y_cum, order)
```
### Cross-Cumulants (2nd, 3rd, and 4th order)
```python
from spectrum import cum2x, cum3x, cum4x, plot_cross_covariance, plot_third_order_cross_cumulant, plot_fourth_order_cross_cumulant
# 2nd order cross-cumulant
ccov = cum2x(x, y, maxlag=20, nsamp=None, overlap=0, flag='biased')
plot_cross_covariance(np.arange(-20, 21), ccov)
# 3rd order cross-cumulant
c3 = cum3x(x, y, z, maxlag=20, nsamp=None, overlap=0, flag='biased', k1=0)
plot_third_order_cross_cumulant(np.arange(-20, 21), c3, k1=0)
# 4th order cross-cumulant
c4 = cum4x(w, x, y, z, maxlag=20, nsamp=None, overlap=0, flag='biased', k1=0, k2=0)
plot_fourth_order_cross_cumulant(np.arange(-20, 21), c4, k1=0, k2=0)
```
## Features
- Estimation of bicoherence and cross-bicoherence
- Direct and indirect methods for bispectrum estimation
- Cross-bispectrum estimation
- Cumulant estimation up to 4th order
- Cross-cumulant estimation up to 4th order
- Plotting functions for all estimations
## Requirements
- Python 3.6+
- NumPy
- SciPy
- Matplotlib
## Contributing
Contributions to the HOSA toolkit are welcome! Please feel free to submit a Pull
Request.
## License
This project is licensed under the MIT License.
## Acknowledgements
This toolkit is based on the Higher Order Spectral Analysis toolkit for MATLAB.
We've adapted and extended it for Python users.
Raw data
{
"_id": null,
"home_page": "https://github.com/synergetics/spectrum",
"name": "higher-spectrum",
"maintainer": null,
"docs_url": null,
"requires_python": ">=3.6",
"maintainer_email": null,
"keywords": "higher order spectrum estimation toolkit signal processing",
"author": "ixaxaar",
"author_email": "root@ixaxaar.in",
"download_url": "https://files.pythonhosted.org/packages/55/51/7280545176addc9638c0e6db270bfbacd715e9803422a4e4251aa1154269/higher_spectrum-0.2.0.tar.gz",
"platform": null,
"description": "# Higher Order Spectral Analysis Toolkit\n\nThis package provides a comprehensive set of tools for higher-order spectral\nanalysis in Python. It includes functions for estimating bicoherence,\nbispectrum, and various orders of cumulants.\n\n## Installation\n\nYou can install the HOSA toolkit using pip:\n\n```bash\npip install higher-spectrum\n```\n\n## Contents\n\n### Bicoherence\n\n```python\nfrom spectrum import bicoherence, plot_bicoherence\n\nbic, waxis = bicoherence(y, nfft=None, window=None, nsamp=None, overlap=None)\nplot_bicoherence(bic, waxis)\n```\n\n\n\n### Cross Bicoherence\n\n```python\nfrom spectrum import bicoherencex, plot_cross_bicoherence\n\nbic, waxis = bicoherencex(w, x, y, nfft=None, window=None, nsamp=None, overlap=None)\nplot_cross_bicoherence(bic, waxis)\n```\n\n\n\n### Bispectrum Direct (using FFT)\n\n```python\nfrom spectrum import bispectrumd, plot_bispectrum\n\nBspec, waxis = bispectrumd(y, nfft=None, window=None, nsamp=None, overlap=None)\nplot_bispectrum(Bspec, waxis)\n```\n\n\n\n### Bispectrum Indirect\n\n```python\nfrom spectrum import bispectrumi, plot_bispectrum_indirect\n\nBspec, waxis = bispectrumi(y, nlag=None, nsamp=None, overlap=None, flag='biased', nfft=None, wind='parzen')\nplot_bispectrum_indirect(Bspec, waxis)\n```\n\n\n\n### Cross Bispectrum (Direct)\n\n```python\nfrom spectrum import bispectrumdx, plot_cross_bispectrum\n\nBspec, waxis = bispectrumdx(x, y, z, nfft=None, window=None, nsamp=None, overlap=None)\nplot_cross_bispectrum(Bspec, waxis)\n```\n\n\n\n### Cumulants (2nd, 3rd, and 4th order)\n\n```python\nfrom spectrum import cumest, plot_cumulant\n\norder = 2 # 2nd order\ny_cum = cumest(y, norder=order, maxlag=20, nsamp=None, overlap=0, flag='biased', k1=0, k2=0)\nplot_cumulant(np.arange(-20, 21), y_cum, order)\n```\n\n### Cross-Cumulants (2nd, 3rd, and 4th order)\n\n```python\nfrom spectrum import cum2x, cum3x, cum4x, plot_cross_covariance, plot_third_order_cross_cumulant, plot_fourth_order_cross_cumulant\n\n# 2nd order cross-cumulant\nccov = cum2x(x, y, maxlag=20, nsamp=None, overlap=0, flag='biased')\nplot_cross_covariance(np.arange(-20, 21), ccov)\n\n# 3rd order cross-cumulant\nc3 = cum3x(x, y, z, maxlag=20, nsamp=None, overlap=0, flag='biased', k1=0)\nplot_third_order_cross_cumulant(np.arange(-20, 21), c3, k1=0)\n\n# 4th order cross-cumulant\nc4 = cum4x(w, x, y, z, maxlag=20, nsamp=None, overlap=0, flag='biased', k1=0, k2=0)\nplot_fourth_order_cross_cumulant(np.arange(-20, 21), c4, k1=0, k2=0)\n```\n\n## Features\n\n- Estimation of bicoherence and cross-bicoherence\n- Direct and indirect methods for bispectrum estimation\n- Cross-bispectrum estimation\n- Cumulant estimation up to 4th order\n- Cross-cumulant estimation up to 4th order\n- Plotting functions for all estimations\n\n## Requirements\n\n- Python 3.6+\n- NumPy\n- SciPy\n- Matplotlib\n\n## Contributing\n\nContributions to the HOSA toolkit are welcome! Please feel free to submit a Pull\nRequest.\n\n## License\n\nThis project is licensed under the MIT License.\n\n## Acknowledgements\n\nThis toolkit is based on the Higher Order Spectral Analysis toolkit for MATLAB.\nWe've adapted and extended it for Python users.\n",
"bugtrack_url": null,
"license": "MIT",
"summary": "Higher Order Spectral Analysis toolkit",
"version": "0.2.0",
"project_urls": {
"Homepage": "https://github.com/synergetics/spectrum"
},
"split_keywords": [
"higher",
"order",
"spectrum",
"estimation",
"toolkit",
"signal",
"processing"
],
"urls": [
{
"comment_text": "",
"digests": {
"blake2b_256": "5f0f395d74df42925f1699c78dcc8a53204b15f170aa9fd967495250c35822df",
"md5": "d07360ccba9ea74f36e7c17cb5e9a2aa",
"sha256": "585fc367c3e046ed0abac986721bf2c640bd22c784150be90d5e85c669065759"
},
"downloads": -1,
"filename": "higher_spectrum-0.2.0-py3-none-any.whl",
"has_sig": false,
"md5_digest": "d07360ccba9ea74f36e7c17cb5e9a2aa",
"packagetype": "bdist_wheel",
"python_version": "py3",
"requires_python": ">=3.6",
"size": 25656,
"upload_time": "2024-10-06T18:28:38",
"upload_time_iso_8601": "2024-10-06T18:28:38.300326Z",
"url": "https://files.pythonhosted.org/packages/5f/0f/395d74df42925f1699c78dcc8a53204b15f170aa9fd967495250c35822df/higher_spectrum-0.2.0-py3-none-any.whl",
"yanked": false,
"yanked_reason": null
},
{
"comment_text": "",
"digests": {
"blake2b_256": "55517280545176addc9638c0e6db270bfbacd715e9803422a4e4251aa1154269",
"md5": "ba1e65b8910bafd2d42c88568a3b0e0c",
"sha256": "f18174cfaf72a1159de1a4d7fb7f553c33afd26306eb3933336c3c06f3c2a3cc"
},
"downloads": -1,
"filename": "higher_spectrum-0.2.0.tar.gz",
"has_sig": false,
"md5_digest": "ba1e65b8910bafd2d42c88568a3b0e0c",
"packagetype": "sdist",
"python_version": "source",
"requires_python": ">=3.6",
"size": 15578,
"upload_time": "2024-10-06T18:28:40",
"upload_time_iso_8601": "2024-10-06T18:28:40.534153Z",
"url": "https://files.pythonhosted.org/packages/55/51/7280545176addc9638c0e6db270bfbacd715e9803422a4e4251aa1154269/higher_spectrum-0.2.0.tar.gz",
"yanked": false,
"yanked_reason": null
}
],
"upload_time": "2024-10-06 18:28:40",
"github": true,
"gitlab": false,
"bitbucket": false,
"codeberg": false,
"github_user": "synergetics",
"github_project": "spectrum",
"travis_ci": false,
"coveralls": false,
"github_actions": false,
"requirements": [
{
"name": "matplotlib",
"specs": [
[
">=",
"3.1.0"
]
]
},
{
"name": "scipy",
"specs": [
[
">=",
"1.4.0"
]
]
},
{
"name": "numpy",
"specs": [
[
">=",
"1.18.0"
]
]
}
],
"lcname": "higher-spectrum"
}
